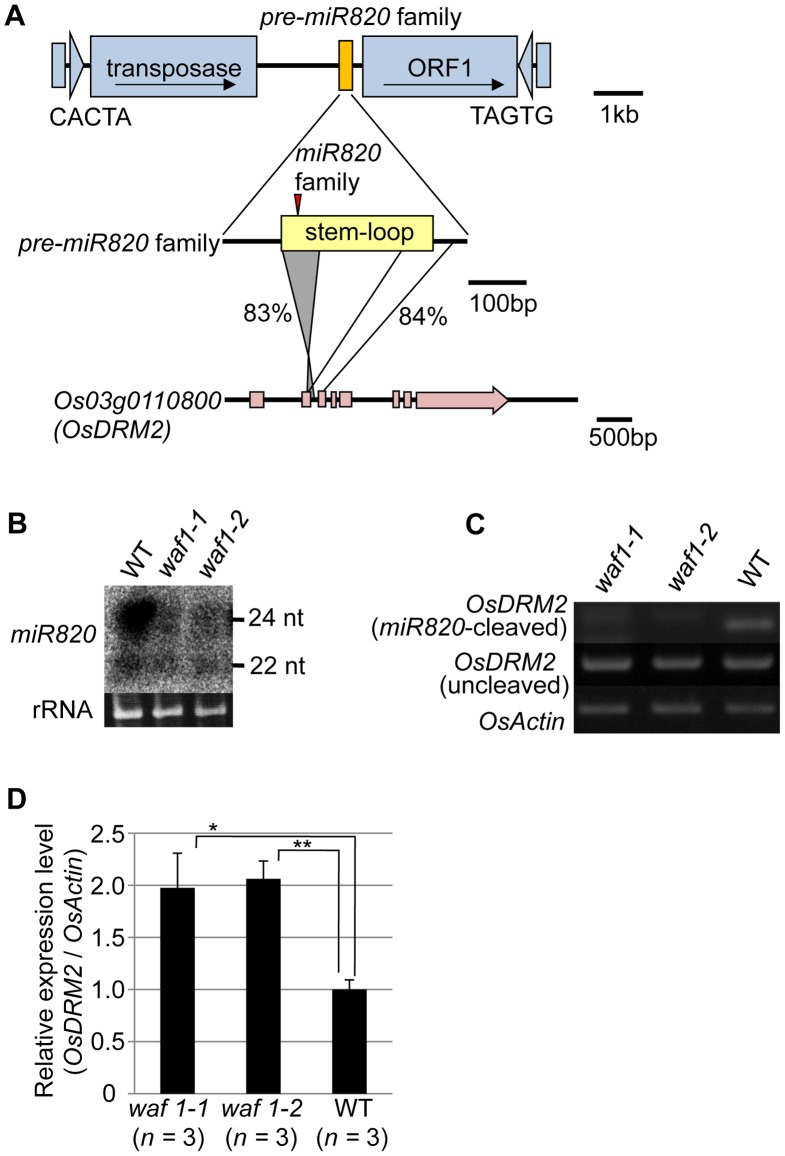
Figure 1. miR820 family members are located within CACTA transposons and target the DNA methyltransferase gene OsDRM2.
(A) The location of miR820 within the CACTA DNA transposon is shown in the first row. The second and third rows indicate the similarity between the sequences of the miR820 precursor (pre-miR820) and Os03g0110800 (OsDRM2). The numbers beside the lines indicate the nucleotide identities between the regions. The red triangle indicates the location of miR820 within the stem-loop region. (B) Northern blot analysis of miR820 expression in the wild-type (WT) and in waf1 mutants. (C) Detection of miR820-cleaved OsDRM2 mRNA by RNA ligation–mediated 5′ RACE (upper panel). The same cDNA templates were used for PCR to amplify OsDRM2 (middle panel) and OsActin (bottom panel) as controls for OsDRM2 expression and RNA integrity. (D) qRT-PCR analysis measuring the expression level of OsDRM2 in waf1-1, waf1-2, and WT. The expression level of the WT was set as 1. Values are means, with bars showing standard errors. Significance was assessed by a two-tailed Student’s t-test; (**) significant at the 1% level; (*) significant at the 5% level. The n value represents the number of mutant or wild-type individuals.