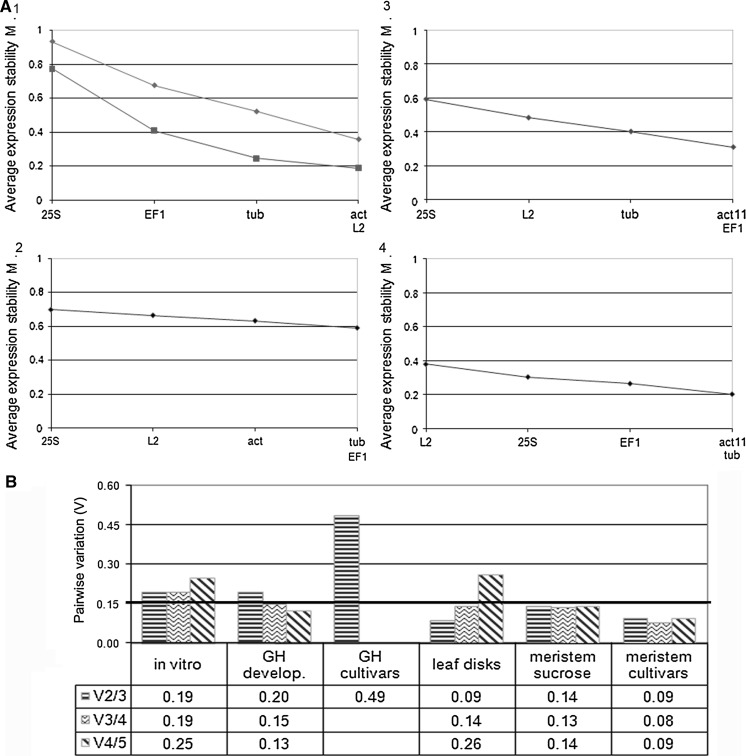
Fig. 2.
Expression stability and variation analyses of the candidate reference genes by geNorm. a Average expression stability (M) and ranking of the candidate reference genes. The lower average expression stability M indicates a more stable expression. Experiments: (1) In vitro (filled diamond), leaf discs (filled square), (2) Greenhouse development, (3) meristem sucrose, (4) meristem varieties. b Pair-wise variation (V) analysis of the candidate reference genes. This analysis was conducted to determine the optimal number of reference genes required for normalization. Six experimental set-ups were included in the analysis: in vitro, greenhouse (GH) development, GH varieties, leaf discs, meristem sucrose, and meristem varieties. A cut-off V-value of 0.15, below which the inclusion of additional reference genes is not required, has been proposed by Vandesompele et al. (2002) and is indicated by a bold line. For the greenhouse development experiment, two reference genes had an M value above 1.5 and only three genes were used to calculate the V pair-wise variation and therefore V3/4 and V4/5 could not be calculated. Abbreviations: ACT11: actin11; ACT: actin; EF1: elongation factor-1α; L2: ribosomal protein L2; 25S: 25S rRNA