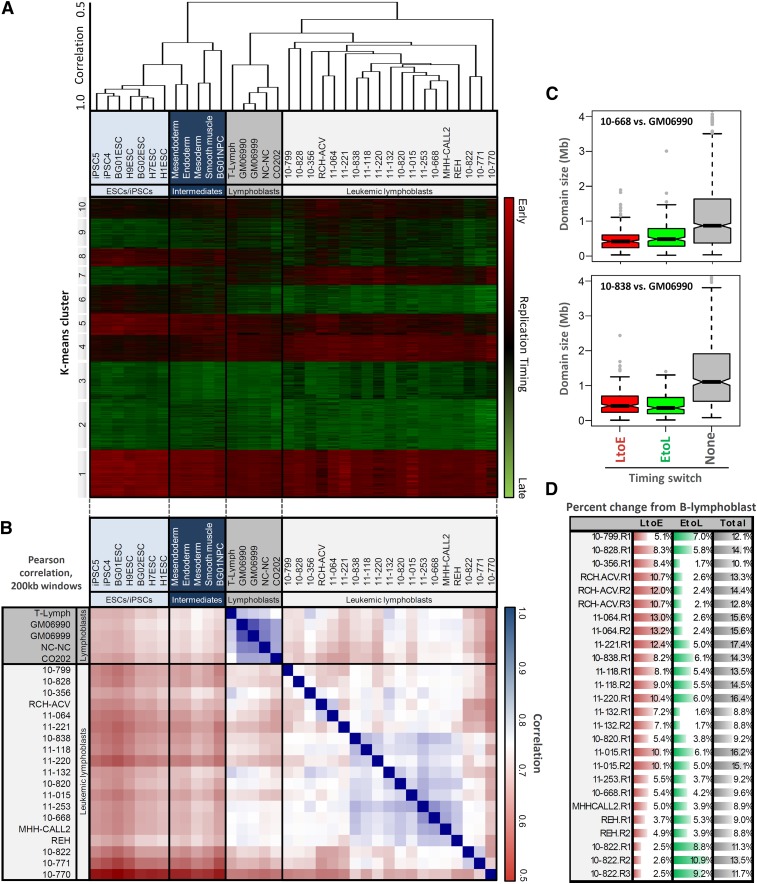
Figure 2.
Leukemic cells show global changes in replication profiles. (A) Hierarchical clustering of genome-wide replication-timing patterns for the four lymphoblastoid B-cell lines and those of other human cell types, showing relatively stable profiles between mature B and T lymphoblasts, and clustering of samples with similar genetic makeup, including T-cell leukemias (10-799/10-828), those with TCF3/PBX1 translocations (11-064/RCH-ACV), and those with mostly normal karyotype (10-838/11-118). (B) Genome-wide correlations between replication-timing data sets used in this study. Correlations between divergent B cells are consistently above 0.9, while those between leukemic samples generally range from 0.60 to 0.85. (C) Domain-wide switches to earlier (L to E) or later (E to L) replication timing occur in units of ∼400–800 kb, smaller than static early or late domains and consistent with developmentally regulated changes in timing. (D) Percentage of the genome with significant timing changes toward earlier (L to E) or later (E to L) replication from the normal B-cell profiles in indicated cell types.