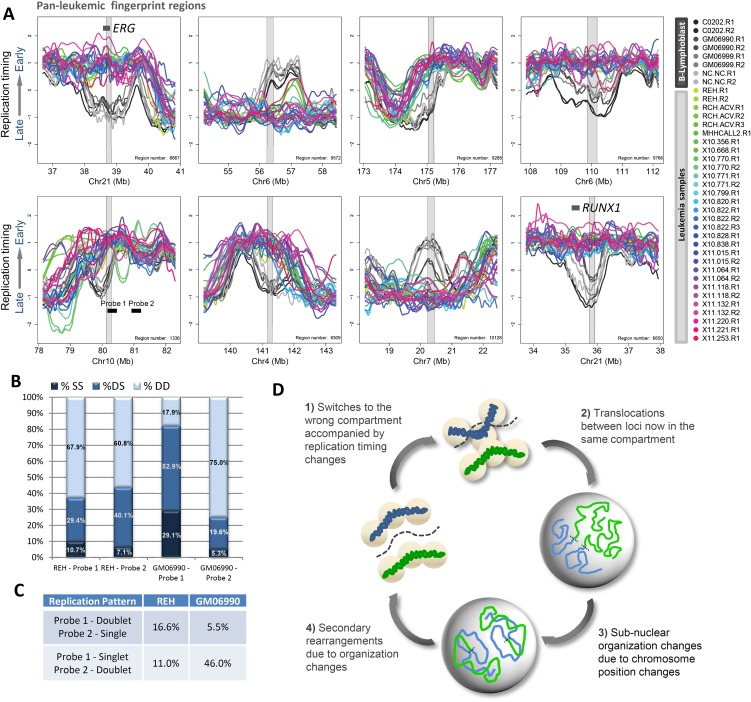
Figure 6.
Pan-leukemic replication-timing changes suggest common early events in leukemogenesis. (A) Example regions from a pan-leukemic fingerprint between all leukemic cells versus B-cell controls. Fingerprint regions are highlighted in gray overlay. (B) Percentages of SS, DS, and DD configurations for each of the FISH probes indicated in A, as probes 1 and 2 were scored as in Figure 5C (192 GM06990 and 516 REH nuclei scored). (C) Quantification of the frequency with which one probe appeared to replicate prior to the other as in Figure 5D (384 GM06990 and 1032 REH chromosomes scored). (D) A prospective model for common early events in leukemogenesis: (1) Loci in late-replicating compartments on the periphery undergo a switch to earlier replication together with a switch to the wrong nuclear compartment, which may be precipitated by loss of anchorage on the periphery or incorporation of accessibility-promoting chromatin factors in early-S phase. (2) Translocations occur between loci that now occupy the same compartment. (3) Large rearrangements between chromosomes disrupt the normal distribution of chromatin in the nucleus, leading to further subnuclear organization changes. (4) Subnuclear organization changes bring together additional loci that would normally not be in contact or share the same compartment, leading to accumulation of additional secondary rearrangements and genome instability.