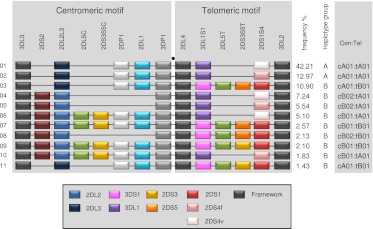
Figure 2.
Common KIR haplotypes. Depicted are the 11 common haplotypes that encompass 94% of Caucasian haplotypes studied. (Right) Haplotype frequencies, A/B group classification, and constituent motifs; for example, cA01 and tA01 denote centromeric-A haplotype 1 and telomeric-A haplotype 1, respectively. Gene and allele content were determined by segregation analysis. (Black spot) Site of the recombination hotspot between 3DP1 and 2DL4. The gene order is based on published sequences of KIR haplotypes that were sequenced in their entirety. Locations of 2DS3S5 may vary on unsequenced haplotypes. The boxed key identifies alleles, variants, and which genes are considered framework. 2DS4v represents the 22-bp frameshift deletion variant, and 2DS4f is the full-length form. 2DL2 and 2DL3 are allelic variants of 2DL2L3, 3DL1 and 3DS1 are variants of 3DL1S1, and 2DS3 and 2DS5 are variants of 2DS3S5. The motif annotation is at the resolution of gene content and does not distinguish allelic variation of individual genes. For example, the haplotype motif tA01 may have either form of 2DS4 (2DS4f or 2DS4v), and tB01 may have either variant of 2DS3S5 (2DS3 or 2DS5). T and C suffixes for 2DL5 (2DL5A or 2DL5B) and 2DS3S5 indicate the telomeric and centromeric versions of the genes, respectively (Gomez-Lozano et al. 2002; Ordonez et al. 2008; Pyo et al. 2010). For simplicity, 2DS1 and 2DS4 are shown at the same site because they showed a mutually exclusive relationship.