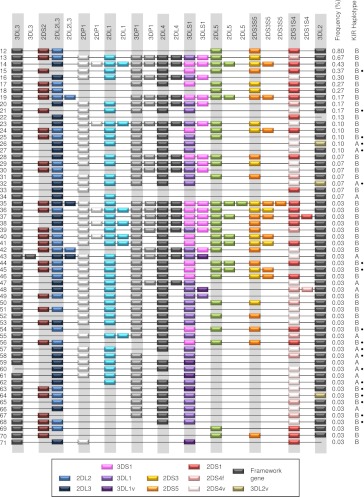
Figure 4.
Unconventional KIR haplotypes and their frequencies. Fifteen altered A haplotypes and 56 B haplotypes all with frequencies <1%. Duplicated genes are not depicted in order, but for simplicity, the paralog has been arbitrarily placed adjacent to the ancestral gene. In some cases, the order of all of the genes has not been determined. Genes suffixed with “v” have parts altered or deleted including fused genes resulting from NAHR or a novel allele. 2DS4v represents the 22-bp frameshift deletion variant, and 2DS4f is the full-length form. 3DL1v on haplotypes 63 and 48 tested negative for exon 4 and on haplotypes 59, 61, 71, and 43 negative for exon 9. 3DL2v on haplotypes 64 and 26 were negative for exon 4 and on haplotype 32 negative for exon 9. Nineteen haplotypes, marked with a black dot, were missing a copy or part of a single locus but otherwise could be the same as standard haplotypes (common arrangements of genes). Apart from haplotypes 15 and 21, which carried a hybrid gene (2DS2/2DS3; 2DS2*005), these marked haplotypes may not reflect true copy number variation because in these cases, we could not rule out the possibility that the gene was present but was erroneously scored as missing due to sequence variation that did not complement our generic primers (see Methods; Supplemental Material). To help resolve novel KIR haplotypes from unphased genotypes of unrelated individuals, we developed a tool for imputing haplotype pairs given observed copy number for each KIR loci. This tool is provided at http://www.bioinformatics.cimr.cam.ac.uk/haplotypes/.