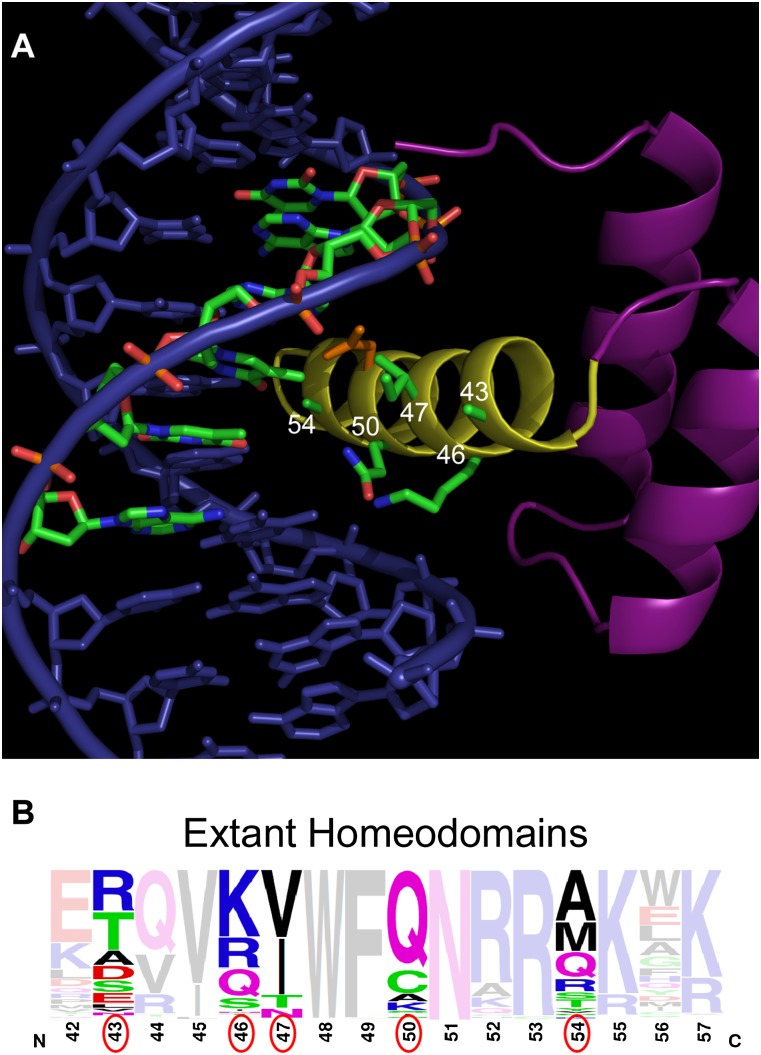
Figure 1.
Structure of the Engrailed HD and distribution of HD recognition residues. (A) Structure of the Engrailed HD–DNA complex (Fraenkel et al. 1998), which serves as the framework for library construction. The numbers (white) on the HD recognition helix (yellow) indicate amino acid positions (green side chains) that were randomized, where the primary strand of the core 6-bp binding site is highlighted (green) to emphasize the proximity of these residues to the 3′ end of the recognition sequence. Asn51 (orange), which is highly conserved within the homeodomain family, is shown for reference. (B) Frequency logo displaying the diversity of residues (the residues randomized in the HD library are circled in red) at various positions in the N51-containing HDs in the genomes of humans, mice, Danio rerio, Caenorhabditis elegans, Drosophila melanogaster, and Saccharomyces cerevisiae.