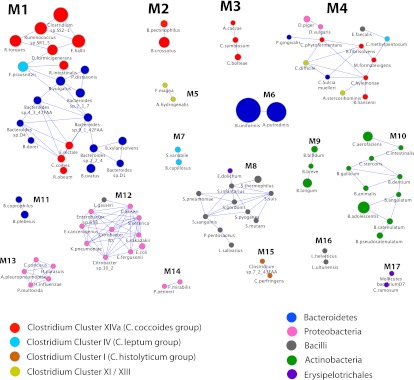
Figure 1.
Co-occurrence network of human gut bacteria, based on a relative abundance matrix previously reported in Qin et al. (2010). The nodes represent species whose genomes have been sequenced. The size of the nodes indicates the average relative abundance across the 124 individuals in the MetaHIT cohort, and the color of the node reflects taxonomic information. Species with significantly positive co-occurrence for any of six measures used (Pearson, Spearman, Kendall, Bray-Curtis, Euclidean, and mutual information) are joined with an edge. Co-occurrence modules are defined as a set/collection of species that are connected among each other (directly or via several steps), but not to any other species in the network, and are labeled with Mx. Species' full names are shown in Supplemental Table S1, and phylogenetic relationships among them as determined with 16S rRNA are shown in Supplemental Fig. S1. For further discussion of this network, see the Supplemental Material.