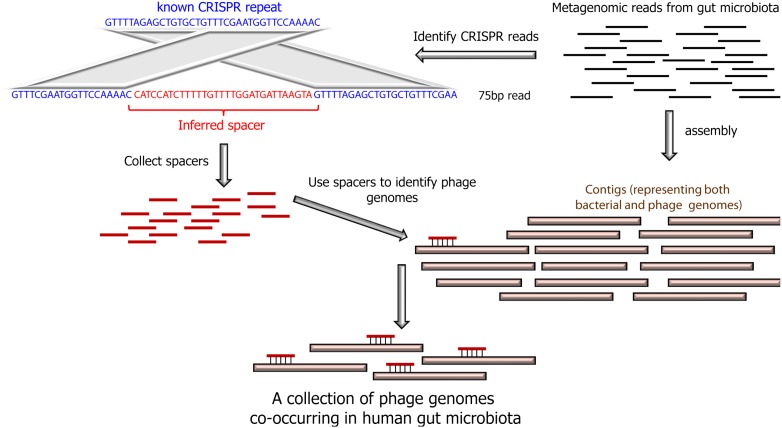
Figure 1.
CRISPR spacers are used as probes to fish out phage genomes. In the MetaHIT metagenomics study (Qin et al. 2010), gut microbes were harvested from feces of 124 individuals, and DNA was sequenced to generate short reads (75 bp). These reads were then assembled into contigs, which mainly represent DNA of gut-residing bacteria, but potentially also contain DNA of phages associated with these bacteria. In the present study, CRISPR spacers were detected by searching for reads that match a known CRISPR repeat on both sides of the read. The spacers detected were then used to probe assembled contigs, and phage and mobile element contigs were identified as those showing high sequence identity with a spacer (but not with the CRISPR repeat).