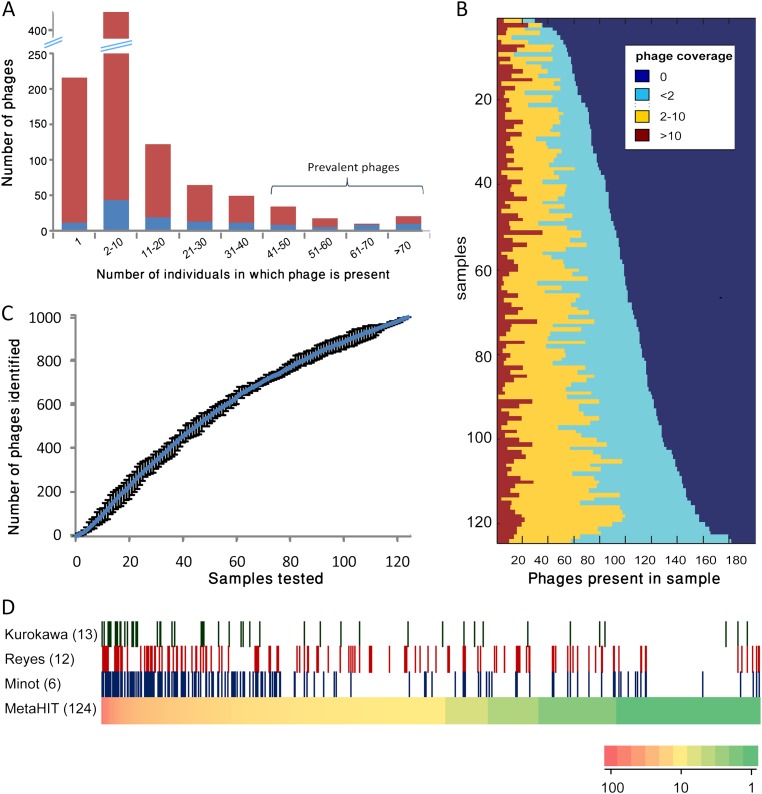
Figure 2.
Phage distribution across individuals and populations. (A) Phage abundance profile. (X-axis) Range of the number of samples in which a phage was present; (y-axis) number of phages whose prevalence is in the range specified. A phage was deemed present if it was significantly covered by metagenomic reads in the sample (Methods). (Blue) Phages shown to integrate into bacterial genome in at least one of the 124 samples; (red) phages with no evidence for integration. (B) Virome profiles of individual samples. Phages are sorted separately in each sample (row) according to their abundance in that sample. (Brown) Highly abundant phages; (yellow) phages of intermediate abundance; (light blue) phages of low abundance. Coverage is measured in reads per kilobase per millions of reads (RPKM). (C) Discovery rate of new phages as a function of number of samples investigated. Samples were added one at a time and in each step spacers were identified. Phage contigs matching to the cumulative set of identified spacers were counted as detected. This analysis was repeated 10 times with random sample order; the charts depict the mean values obtained over the 10 iterations, and bars demarcate maximum and minimum values. (D) Evidence for presence of phages identified in this study (based on MetaHIT data) (Qin et al. 2010) in VLP data derived from American individuals (Reyes et al. 2010; Minot et al. 2011), and in gut metagenomic data derived from Japanese individuals (Kurokawa et al. 2007). (Bottom row) 991 phage contigs as bars sorted by prevalence in the MetaHIT population (heatmap). In each of the top three rows, a bar is colored if the phage is significantly covered by reads from the data set listed. The number of individuals in each data set is in parentheses.