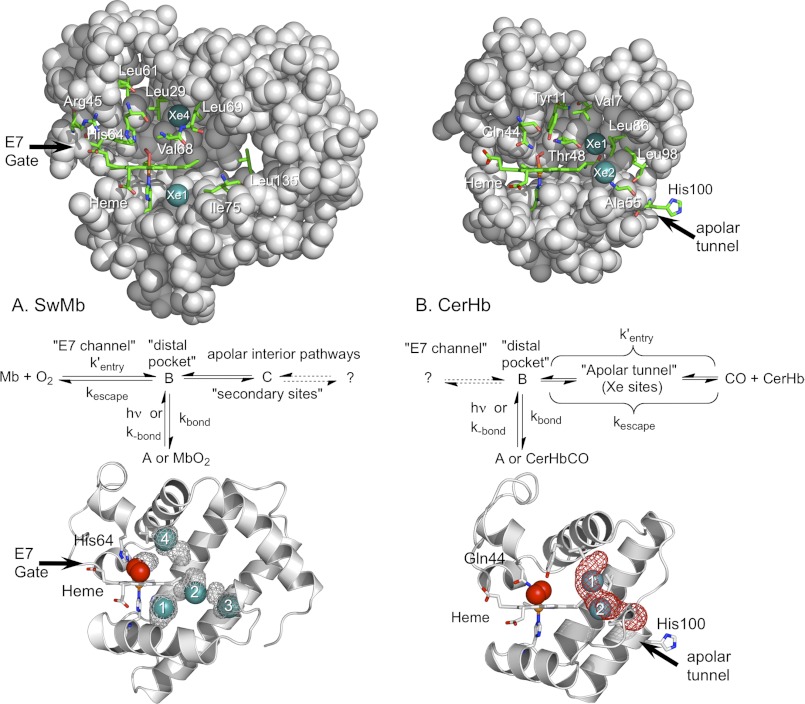
FIGURE 1.
Side views of WT CerHbO2 and SwMbO2. In both globins the molecules are oriented so that the planar heme group is viewed edge-on, its propionates are facing left and the “distal pocket” is above the heme. Portions of the protein matrix are removed so the interior view is clear. Xe atoms are depicted as teal spheres; key amino acids and the heme prosthetic groups are indicated by sticks with carbon in green, oxygen in red, and nitrogen in blue; the remainder of the protein structure is represented as gray spheres. A, upper panel, SwMbO2 with most of the E-helix removed (PDB code 2MGM) and Xe1 and Xe4 inserted into positions obtained from the metMb structure determined at high Xe pressures (PDB code 2W6W). Key distal pocket residues include Leu-29(B10), His-64(E7), Val-68(E11), and Phe-43(CD1). Additional mutants constructed in this study include mutations made at residues Arg-45(CD3), Leu-61(E4), Leu-69(E12), Ile-75(E18), and Leu-135(H12). Middle panel, mechanism for ligand binding to Mb based on Scott et al. (13, 38). Lower panel, cavity surfaces (gray mesh) in SwMbO2 (PDB code 2MGM), obtained using the surface function in PyMOL, are compared with the Xe atom positions found in metMb under high Xe pressures (PDB code 2W6W). As described in Pesce et al. (39), no apolar tunnels were found in SwMb using Surfnet (64). We also found no tunnel in SwMb using MOLE online 2.0 (65) using the default settings. The cavities found using PyMOL in SwMbO2 are not connected. Xe1–Xe4 atoms were inserted into the structure based on their positions in 2W6W. B, upper panel, WT CerHbO2 (PDB code 1KR7) with most of the E helix removed and bound Xe atoms inserted at their locations in L86A CerHbO2 equilibrated with Xe gas (PDB code 2XKH). Key distal pocket and apolar tunnel residues include Val-7(B6), Tyr-11(B10), Gln-44(E7), Thr-48(E11), Ala-55(E18), Ala-82(G8), Leu-86(G12), Leu-98(H9), and His-100(H11) in WT CerHbO2. Middle panel, mechanism for ligand binding to CerHb taken from Salter et al. (42) and Pesce et al. (43). Lower panel, ribbons drawing for WT CerHbO2 (1KR7) with an apolar tunnel (red mesh) computed using MOLE online 2.0 (65) with a probe radius of 3 and 1.25 Å interior threshold (default settings). Xe atoms were inserted based on their positions in PDB code 2XKH.