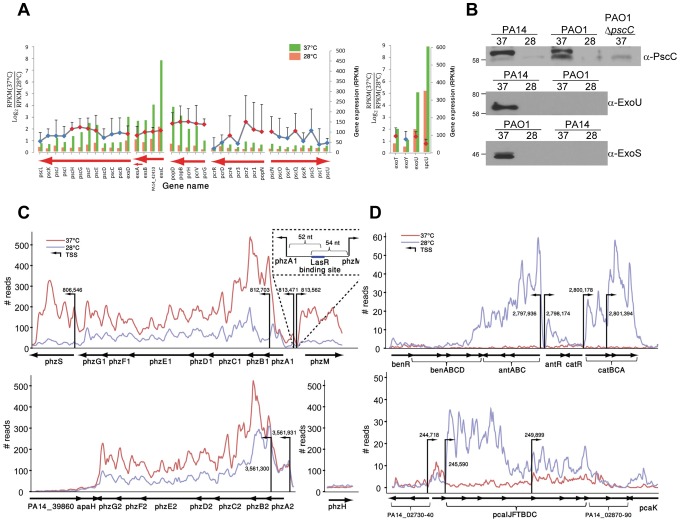
Figure 2. Temperature-regulated expression of selected virulence factors.
(A) The T3SS of P. aeruginosa is induced at 37°C. Transcript levels of the genes encoding the T3SS components; its regulators; secreted effectors ExoT, Y, U; and the chaperone SpcU. Normalized gene expression [78] (Materials and Methods) for each gene in the two growth temperatures is represented by bars for each gene (green and red, 37°C and 28°C, respectively). The log2-ratio for 37°C and 28°C expression is represented by a diamond for each gene [red diamonds represent a statistically significant difference (χ2, p<0.05) in expression for two replicates while genes marked with blue diamonds did not pass the significance threshold]. Genes connected by a line between their representing diamonds are part of the same transcriptional unit (TU). Error bars represent a single standard deviation from the mean. Horizontal red arrows represent TUs and their strand-orientation (right and left, forward and reverse strand, respectively). (B) Analyses of expression of the secretin and exoproteins of the T3SS in strains PA14 and PAO1. Cellular (top panel) and secreted (middle and lower panel) proteins from cultures growing at 37 or 28°C (labeled 37 or 28 respectively) were separated by SDS-PAGE followed by Western immunoblotting using antibodies against PscC (α-PscC), ExoU (α-ExoU), or ExoS (α-ExoS). The migration of molecular protein standards are indicated in kilodaltons. The common band on all blots probed with the α-PscC antibody represents a cross-reactive protein. (C) Expression of genes for the phenazine biosynthesis operons. The normalized number of sequencing reads mapped to the genes (black arrows) in the phenazine biosynthesis operons is shown in 37°C and 28°C (red and blue lines respectively). The TSSs that were detected in the operons are shown as black vertical lines (arrowheads representing the strand of the transcription, right and left, forward and reverse, respectively; numbers represent the genomic position of the TSS). A palindromic LasR binding site was identified 52 nt upstream to the TSS of the phzA1-G1 operon and 54 nt downstream of the TSS of phzM. (D) Expression of genes for enzymes in pathways that contribute precursors to the biosynthesis of phenazines.