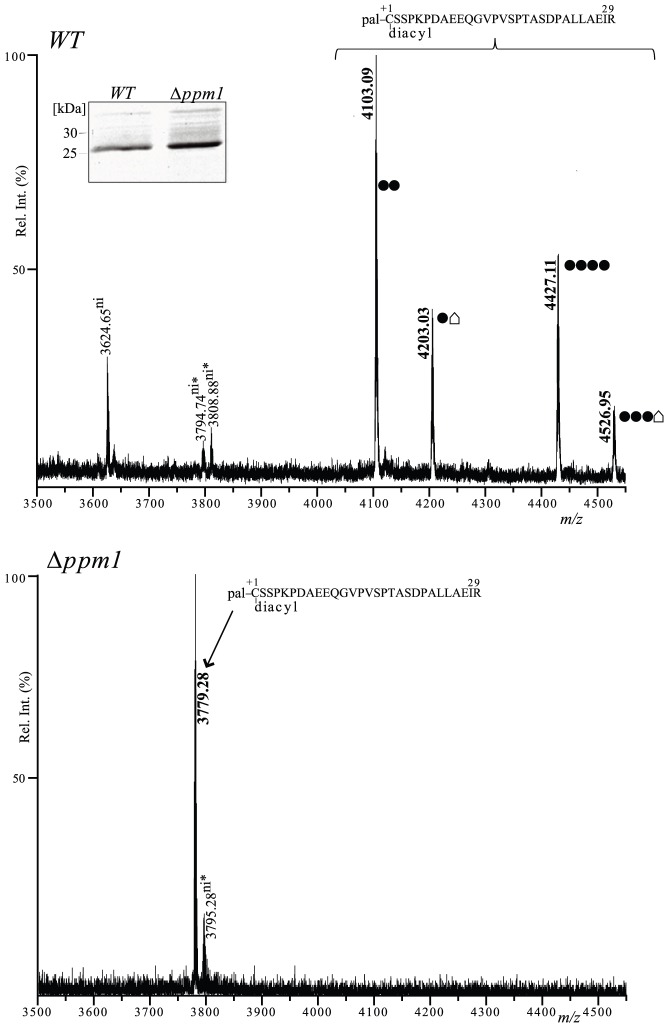
Figure 4. Cg-Ppm1 is required for LppX glycosylation.
Comparison of MALDI PMF profiles of LppX protein purified from C. glutamicum wild-type and Δppm1 strains. The m/z 3500–4550 region of the mass spectra of LppX tryptic peptides after DDM/CHCl3-CH3OH treatment is shown and significant monoisotopic [M+H]+1 peaks are annotated. Upper panel: m/z peaks correspond to glycosylated and triacylated LppX1–29 peptides are indicated in bold (m/z 4103.09, 4203.03, 4427.11 and 4526.95). Three peaks (m/z 3624.65, 3794.74 and 3808.88) were detected but not identified. Bottom panel: the m/z peak corresponding to the non-glycosylated triacylated LppX1–29 peptide was specifically observed in the Δppm1mutant (m/z 3779.28, in bold). The m/z 3795.28 peak was not identified. • = 1 hexose (Δm = 162 Da). • = unknown modification (Δm = 262 Da). “ni” means not identified and asterisks indicate that m/z assignments are not very accurate. Inset: SDS PAGE of the purified LppX proteins from the wild type and the Δppm1 strains.