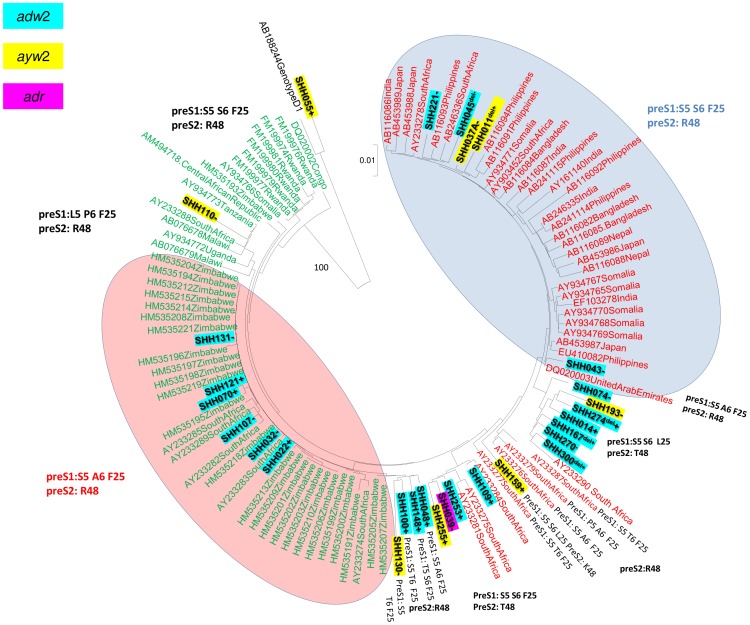
Figure 1. Phylogenetic relationship of complete pre-S1/pre-S2/S sequences (nt 2854–835 from the EcoRI site, numbering according to GenBank accession #AY233274) of 29 HBV isolates from HIV infected particpants [isolate number in bold, +: HBsAg+ve, −:HBsAg−ve, del: deletion mutant] to sequences of other African (green) and “Asian” (red) subgenotype A1 HBV isolates obtained from GenBank established using neighbour-joining.
Bootstrap statistical analysis was performed using 1000 data sets and the numbers on the nodes indicate the percentage of occurrences. Each sequence obtained from GenBank is designated by its accession number and its country of origin. The characteristic amino acids in S open reading frame are indicated next to the sequences or relevant clades.