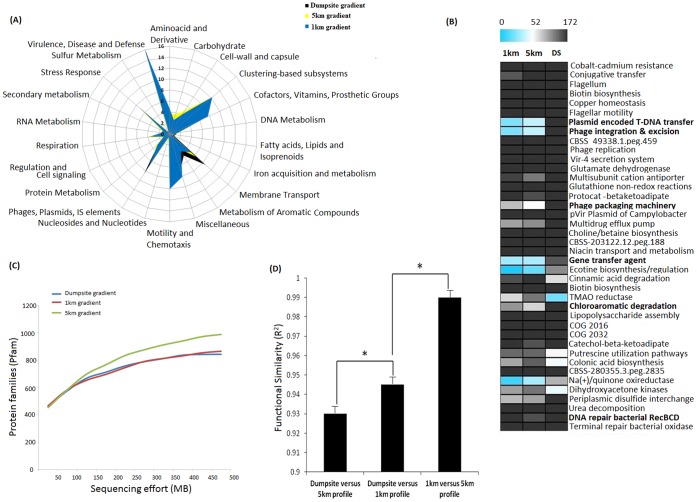
Figure 2. Functional traits of the studied metagenomes.
(A) Cellular processes enriched over increasing HCH contamination. Metagenomic reads were compared against the COG database and relative percentage (y-axis) for each category (x-axis) was calculated. (B) Heat map showing the relative abundance of top 50 subsystems enriched over increasing HCH concentrations (percentage cut-off = 0.8%, standard deviation cut-off = 0.4%). (C) Rarefaction analysis performed on unique protein families (Pfam) sampled across three HCH gradients. (D) Comparison of functional categories similarity between metagenome gradient pairs. KEGG enzyme profile of each metagenome was compared. Asterisks indicate significant differences (Two sided Fishers exact test with Bonferroni multiple test correction, P<0.01). ABBREVATIONS: (1) DS = dumpsite gradient, 1 km = 1 km gradient and 5 km = 5 km gradient.