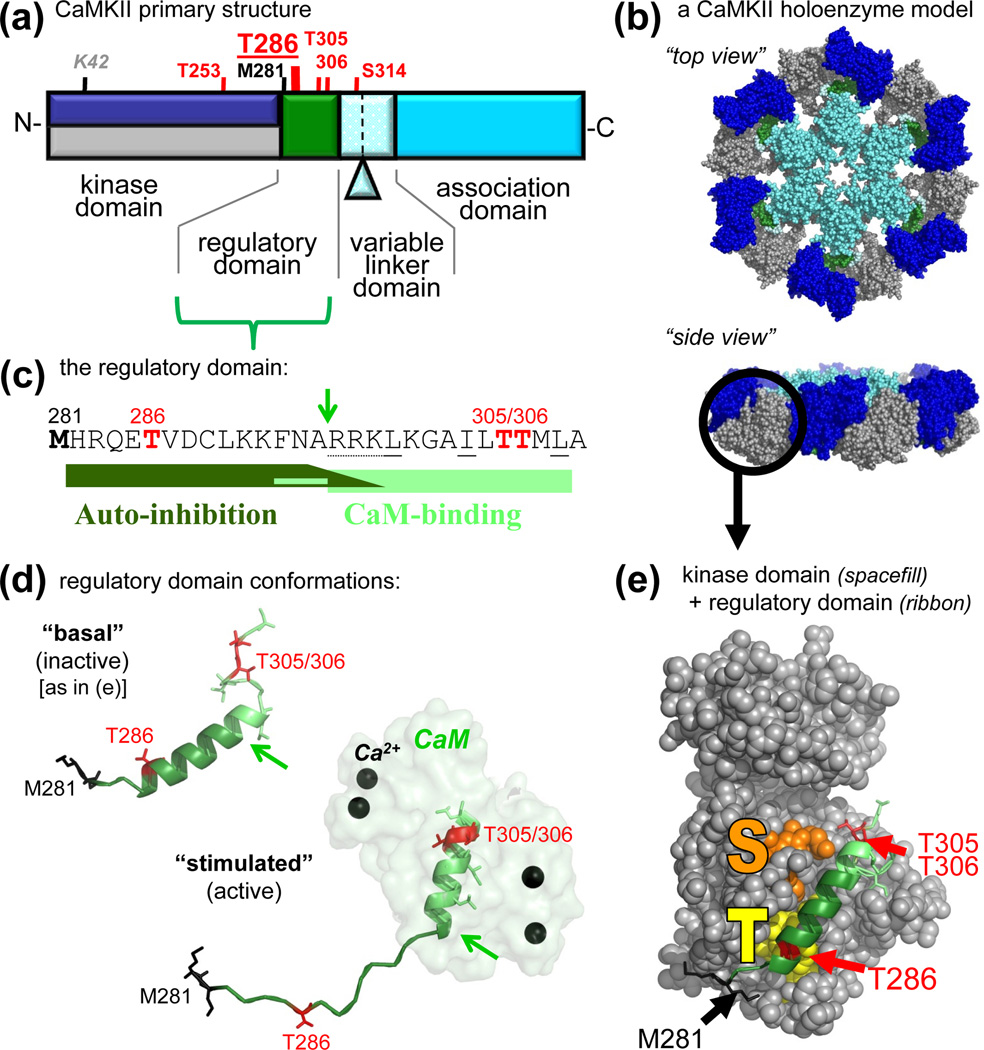
Figure 1.
CaMKII structure and regulation. (a) Primary structure of CaMKIIα, with an N-terminal kinase domain, followed by a CaM-binding regulatory domain, a variable linker domain (subject to alternative splicing, with the splice insert of αB indicated), and a C-terminal association domain. Phosphorylation sites (red) and other important residues are indicated (see also Table 1). The other isoforms (β, γ,δ) contain an additional Thr after the third residues, and the numbers of their homologous positions are accordingly higher by one. These other isoforms also show more extensive alternative splicing in the variable domain, which is typically longer. (b) A model of a 12meric CaMKII holoenzyme (in two different view angles), based on several partial crystal structures [44, 46, 59], with the association domains forming a central hub [44, 46], and the kinases domain [59] radiating outward. The 12meric nature, the particle size, and the principle arrangement is supported by EM studies [44, 45]; however, the precise positioning of the kinase domains in this holoenzyme conformation is unclear (but compare Fig. 4). The reconstruction was kindly provided by Drs. Chao and Kuriyan, and the image is adapted, with permission, from [35]. (c) Sequence of the regulatory domain. The autoinhibitory and CaM-binding regions are indicated. The arrow indicates the N-terminus of the core CaM-binding region (R296; also indicated by arrows in panel d); Ca2+/CaM trapping on the T286-phosphorylated form of the kinase also involves F293 [2]. Autoinhibition is maintained in a kinase 1-N294 truncation, but additional C-terminal residues participate in the substrate site block [2] (see also panel e). The oxidation (black) and phosphorylation (red) sites that generate autonomous activity (T286) [2, 29, 30] or inhibit CaM-binding (T306/306) [2, 70–72] are marked. (d) The regulatory domain in the basal (inactive) and stimulated (active) conformation, illustrating a structural transition of the different regions upon Ca2+/CaM binding [32]. The positioning relative to the kinase domain is known for the basal state (see panel e) but unclear for the active states. The region shown here for CaMKIIδ is identical in CaMKIIα. (e) Crystal structure of human CaMKIIδ in its basal state, with the regulatory domain (ribbon) held in place in part by interactions with the T286 binding T-site (yellow) and blocking access to the substrate binding S-site (orange). The structural representations in panels d and e are based on Protein Data Bank (PDB) files 2VN9 and 2WEL [32].