FIGURE 1.
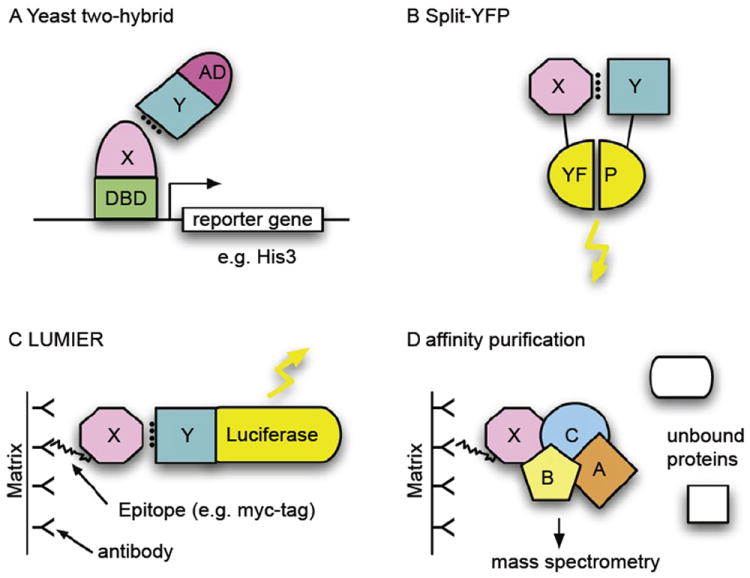
Selected methods for the study of protein–protein interactions. (A) The yeast two-hybrid (Y2H) system is based on the levels of two fusion proteins (typically in yeast but any cell type can be used). One of the proteins contains a DNA-binding domain (DBD), which can bind to the promoter of a reporter gene (here: HIS3), and a second protein X, the bait. The second fusion protein consists of a transcription-activation domain (AD) and a second protein, Y. If proteins X and Y interact, a transcription factor is formed and the reporter gene is activated. In this case, that means that the cell can grow on histidine-free medium. A yeast colony growing on such medium thus indicates an interaction of the two inserted proteins. (B) Protein complementation assay (PCA), e.g., split-YFP. As in the Y2H assay, two interacting proteins bring together two protein fragments that are inactive when separate but active when in close proximity. Here, fragments of yellow-fluorescent protein (YFP) reassociate and fluoresce when reassembled. Other fluorescent proteins, such as the green fluorescent protein (GFP), have been used in a similar way. (C) LUMIER (LUMInescence-based mammalian intERactome). Two fusion proteins are purified by means of an epitope tag (here: FLAG tag), usually on an antibody-coated matrix. Interactions between X and Y can be detected using Luciferase, whose gene is fused to Y and which emits light when luciferin is added. (D) Affinity purification. Protein complexes can be purified from cellular lysates using an affinity epitope, as in (C) with a FLAG tag. Components of the complex can then be identified using mass spectrometry, using the unique mass of peptides when the protein is digested by trypsin.
