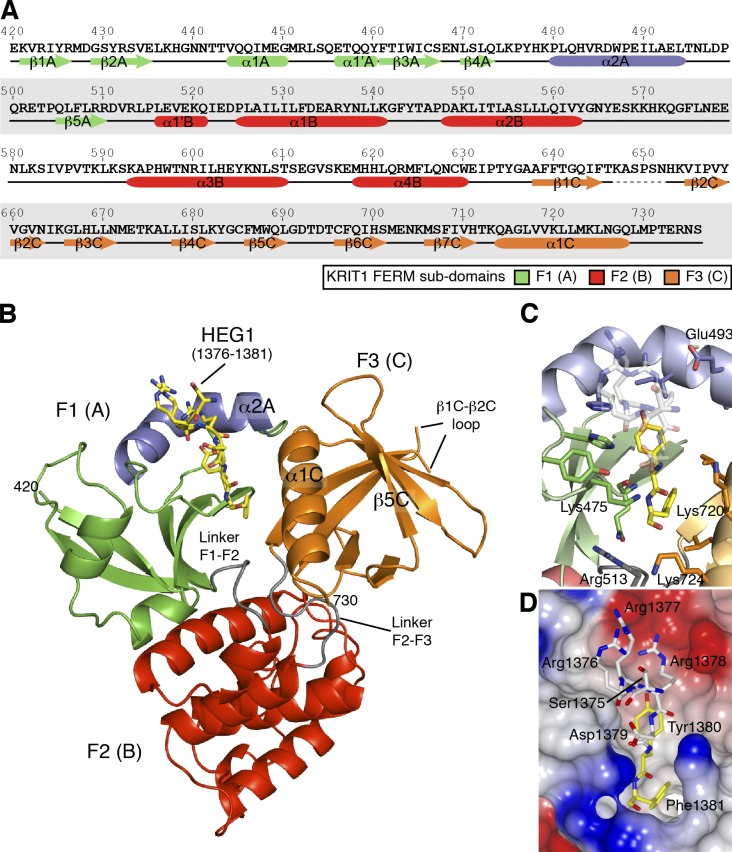
Figure 2.
Crystal structure of the KRIT1 FERM domain bound to the HEG1 cytoplasmic tail peptide reveals a new mode of binding. (A) Sequence of human KRIT1 (UniProt O00522) FERM domain with secondary structure elements shown below the sequence. (B) View of the KRIT1 FERM domain bound to the HEG1 peptide. The HEG1 peptide is shown in yellow. The KRIT1 FERM domain consists of three subdomains: F1 (green and blue), F2, and F3. The linkers F1–F2 (residues 511–515) and F2–F3 (residues 631–637) are colored in gray, and the new features of the F1 domain that are not present in ubiquitin and radixin are shown in blue, i.e., helix α2A (residues 480–494). (C) The HEG1 binding pocket at the KRIT1 F1 and F3 interface. The interaction is mainly formed by hydrogen bonds from residues in loop β4A–α2A and hydrophobic residues from helix α2A and α1C. (D) Surface electrostatic potentials of the KRIT1 FERM domain with the HEG1 peptide shown as a stick model. The HEG1 residues buried in the hydrophobic pocket are shown in yellow, whereas the charged residues exiting the pocket are shown in white.