Transcriptional regulation by Rb–E2F and posttranscriptional regulation by microRNAs control the expression of cell cycle and DNA replication genes and restrict cellular proliferation.
Abstract
The cancer-associated loss of microRNA (miRNA) expression leads to a proliferative advantage and aggressive behavior through largely unknown mechanisms. Here, we exploit a model system that recapitulates physiological terminal differentiation and its reversal upon oncogene expression to analyze coordinated mRNA/miRNA responses. The cell cycle reentry of myotubes, forced by the E1A oncogene, was associated with a pattern of mRNA/miRNA modulation that was largely reciprocal to that induced during the differentiation of myoblasts into myotubes. The E1A-induced mRNA response was preponderantly Retinoblastoma protein (Rb)-dependent. Conversely, the miRNA response was mostly Rb-independent and exerted through tissue-specific factors and Myc. A subset of these miRNAs (miR-1, miR-34, miR-22, miR-365, miR-29, miR-145, and Let-7) was shown to coordinately target Rb-dependent cell cycle and DNA replication mRNAs. Thus, a dual level of regulation—transcriptional regulation via Rb–E2F and posttranscriptional regulation via miRNAs—confers robustness to cell cycle control and provides a molecular basis to understand the role of miRNA subversion in cancer.
Introduction
MicroRNAs (miRNAs) are an evolutionarily conserved class of small (18–22 nucleotides) noncoding RNAs that have emerged in recent years as pivotal regulators of a variety of cellular processes, including differentiation, growth control, and apoptosis (Bushati and Cohen, 2007; Bartel, 2009). miRNAs were originally discovered in worms as developmental regulators (i.e., lin-4/let-7; Wightman et al., 1993; Reinhart et al., 2000), but they have since been found to have a tissue- or lineage-specific pattern of expression in higher organisms as well, underlying their direct involvement in tissue differentiation and homeostasis in multicellular organisms (Thomson et al., 2004; Landgraf et al., 2007). miRNAs regulate gene expression at a posttranscriptional level, with each miRNA being able to target several mRNA species (Bartel, 2009). Thus, these small molecules are able to establish complex networks of interactions to coordinate cellular responses (Stark et al., 2005; Krol et al., 2010; Mestdagh et al., 2011; Bissels et al., 2011). Deregulation of miRNA expression has been associated with multiple diseases including cancer (Calin and Croce, 2006; Kloosterman and Plasterk, 2006), where a global reduction of miRNAs has been often observed as a general trait (Lu et al., 2005; Chang et al., 2008; Ozen et al., 2008; Kumar et al., 2009). Although loss of miRNAs in cancer (and in particular loss of differentiation-associated miRNAs [DA-miRNAs]) could be consequential to the undifferentiated state of advanced tumors, several molecular and genetic lines of evidence suggest that the reduction in miRNA levels is functional to cell transformation and associated with an aggressive disease phenotype (Kumar et al., 2007; Merritt et al., 2008; Martello et al., 2010). To date, however, the phenotypic advantages of reduced miRNA expression to tumor cell proliferation and progression remain poorly characterized.
Given the tight relationship between proliferation and differentiation in physiology and in cancer (Croce, 2008; Hanahan and Weinberg, 2011), we hypothesized that DA-miRNAs can inhibit proliferation, thus cooperating with known regulatory gene expression pathways that, under physiological conditions, keep cell proliferation tightly reined in. To investigate the relationship between cancer-promoting proliferative pathways and DA-miRNAs, we used a cell model consisting of committed progenitors (myoblasts) induced to terminally differentiate into myotubes (terminally differentiated [TD] cells) and then forced to reenter the cell cycle by the E1A adenoviral oncogene (Crescenzi et al., 1995). The adenoviral E1A protein behaves as a potent oncogene that can revert the tightly controlled state of TD cells by inducing proliferation and dedifferentiation pathways, thus mimicking common alterations in human cancer (Nicassio et al., 2005; Bianchi et al., 2007). We used this model to explore how mRNAs and miRNA components are regulated and how they are integrated to effect specific phenotypes.
Our results indicate that, during terminal differentiation, the expression of cell cycle and DNA replication genes is repressed by the Retinoblastoma protein (Rb) tumor suppressor pathway. This repression is released by the E1A oncogene during cell cycle reentry. Additionally, we show that a significant fraction of Rb-dependent genes are also regulated posttranscriptionally in an Rb-independent manner, via a subset of DA-miRNAs. This dual level of regulation of Rb-dependent messenger RNAs (directly via the Rb–E2F axis and indirectly via Rb-independent miRNAs) reinforces the postmitotic block that occurs during terminal differentiation, and provides a safety mechanism to protect against oncogene-induced proliferation.
Results
E1A induces reexpression of proliferative genes through its interaction with Rb
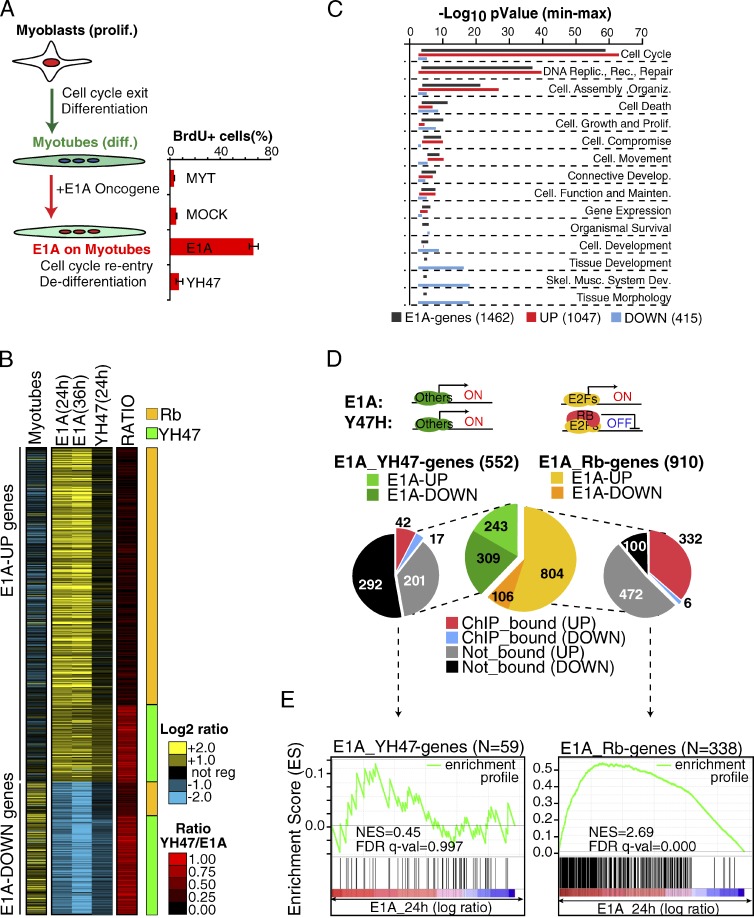
Proliferating C2C12 myoblasts were induced to terminally differentiate (TD) into myotubes and were then infected with a recombinant adenovirus expressing E1A (Ad-E1A), which induces cell cycle reentry and dedifferentiation (Fig. 1 A). We analyzed the gene expression program induced during differentiation of myoblasts into myotubes (Fig. 1 B, Myotubes), and after E1A expression in myotubes (E1A vs. Mock infected myotubes; Fig. 1 B, “E1A”), using the Affymetrix GeneChip 430 platform. We found 1,462 genes significantly regulated (corrected P ≤ 0.05) by E1A during cell cycle reentry (henceforth the “E1A mRNA signature”), of which 1,047 (71.6%) were up-regulated (E1A-UP genes) and 415 (28.4%) were down-regulated (E1A-DOWN genes). We observed that E1A largely reverts the transcriptional program associated with myotube differentiation (911 oppositely regulated genes, P < 0.0001; Fig. 1 B). Functional classification analysis revealed that, although E1A-UP genes are significantly enriched in proliferative functions (cell cycle and DNA replication), E1A-DOWN genes are primarily involved in development, differentiation, and skeletal muscle functions (Fig. 1 C and Table S2).
Figure 1.
E1A-TD mRNA signature (GEX-C2C12). (A) Scheme of the E1A-TD model. The bar graph shows the quantification of cell-cycle reentry induced by E1A or the YH47 mutant by BrdU incorporation assay in two independent experiments (error bars indicate mean and standard deviations). (B) The heat map shows the 1,462 E1A-regulated genes at each time point. Gene expression changes during differentiation (“Myotubes”) and after the expression of the E1A YH47/dl928 mutant (“YH47 24 h”) are also shown. The ratio between the YH47 mutant and wt-E1A regulation was used to distinguish Rb genes (Rb, in orange) and YH47 genes (YH47, in green). (C) Functional analysis of the E1A-TD mRNA signature. Bars represent the Min-Max p-value (−log10 scale) for the most significant (P < 0.001) functional classes calculated over all E1A genes, or over up- and down-regulated gene classes separately. (D) A series of pie charts summarize the E1A-TD mRNA signature. The central chart illustrates the transcriptional class of the signature according to the YH47/E1A ratio. Side charts show the overlap with Rb ChIP-bound promoters by meta-analysis. (E) GSEA was used to correlate the transcriptional classes of E1A-TD mRNA signature (Rb genes and YH47 genes) with Rb-bound promoters by ChIP-seq (Chicas et al., 2010). Significance (FDR) and NES are shown.
E1A interferes with the function of the tumor suppressor protein Rb and that of the related proteins p107 and p130 (collectively referred to as “pocket proteins”; Lipinski and Jacks, 1999). To understand the impact of this pathway on the E1A-induced transcriptional reprogramming of TD-C2C12 cells, we took advantage of the E1A mutant YH47/dl928 (henceforth YH47), which cannot bind to pocket proteins and which is unable to force TD cells to reenter the cell cycle (Fig. 1 A; Tiainen et al., 1996; Nicassio et al., 2005). By comparing the gene expression profiles of TD-C2C12 cells infected with E1A or YH47, we observed that the regulation of the majority of the E1A genes was through binding to pocket proteins (910 of 1,462 genes, 62%; Fig. 1, B and D; we refer to these genes as “Rb genes”). However, although most of the E1A-UP genes were Rb-regulated (804 of 1,047, 77%, P < 0.0001), the regulation of the majority of the E1A-DOWN genes was Rb-independent (309 of 415, 76%, P < 0.0001). By unsupervised TRANSFAC analysis, we found a significant enrichment for E2F family binding sites at gene promoters of E1A-UP genes (in particular the Rb-dependent ones; Table S3). Instead, the promoters of YH47-induced genes (genes induced by E1A and also by YH47, i.e., Rb-independent) were devoid of such binding sites.
We compared our Rb gene list with genes in a published genome-wide analysis of Rb-bound promoters identified through a chromatin immunoprecipitation (ChIP) sequencing approach (Chicas et al., 2010). We observed a significant overlap (P < 0.0001; Fig. 1 D) and a very high correlation (normalized enrichment score [NES] = 2.69 by gene set enrichment analysis [GSEA]; Fig. 1 E) between the Rb genes (and in particular those up-regulated by E1A) and Rb-ChIP bound promoters. Comparable results were obtained in a related cellular model, which consists of TD primary muscle satellite cells (MSCs) induced to reenter the cell cycle by E1A expression (Fig. S1). In this latter model, we identified Rb-dependent genes through a different independent approach, namely the genetic depletion of a floxed Rb locus by Cre recombinase expression (Fig. S1 A).
Collectively, these data establish that the up-regulation of proliferative genes by the interaction with the Rb–E2F pathway is the major component of the E1A-induced transcriptional (mRNA) response during cell-cycle reentry of TD cells.
E1A down-regulates DA-miRNAs through an Rb-independent transcriptional mechanism
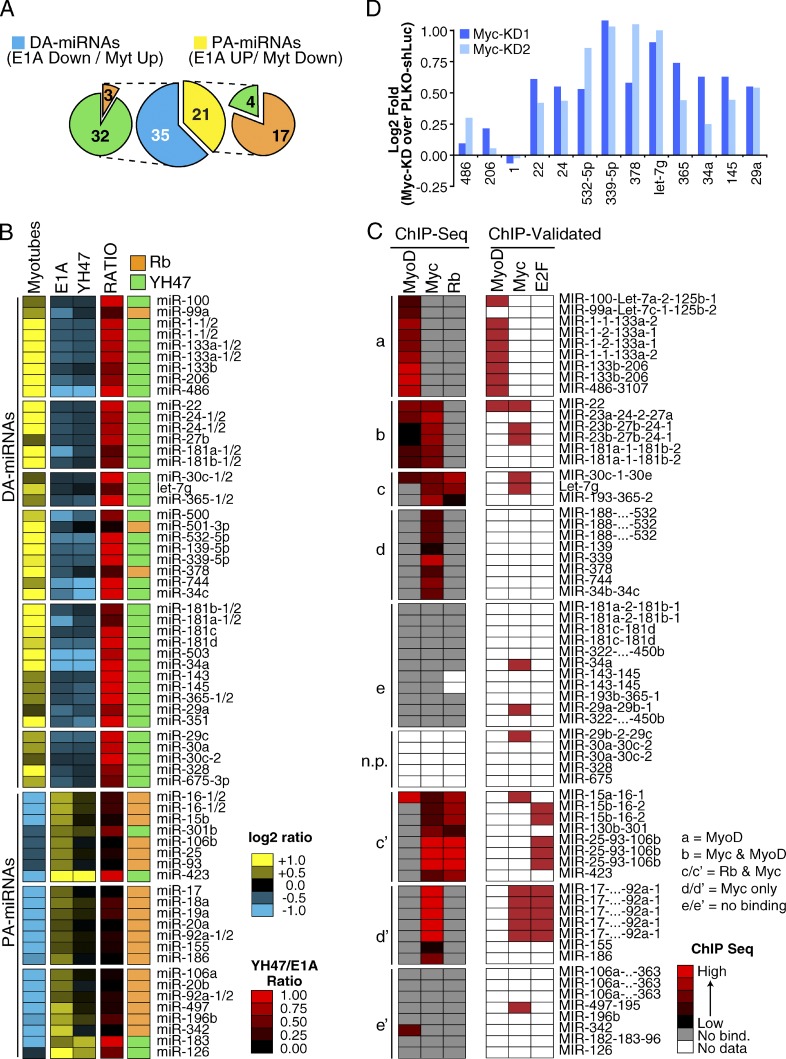
We analyzed miRNA expression upon terminal differentiation of C2C12 cells and during their subsequent E1A-induced cell cycle reentry. By measuring the expression of 335 murine miRNAs by RT-qPCR (see Materials and methods for details), we found a total of 56 regulated miRNAs, which were distinguishable according to their class of regulation: DA-miRNAs consist of miRNAs induced during differentiation and repressed by E1A expression (n = 35); proliferation-associated miRNAs (PA-miRNAs) instead consist of miRNAs repressed during differentiation and induced upon cell cycle reentry by E1A (n = 21; Fig. 2, A and B; and Table S4). We refer to this set of 56 miRNAs as the E1A-TD miRNA signature. We obtained similar results in MSCs (54 DA-miRNAs and 21 PA-miRNAs in MSCs; Fig. S2, A and B; and Table S4), and, in fact, we observed a significant overlap between the E1A-TD signatures in the two model systems (28/35 DA-miRNAs and 14/21 PA-miRNAs, P < 0.0001). E1A expression, therefore, has a greater effect on miRNA repression than on miRNA induction, something that is particularly evident in the MSC model, and is reminiscent of the wide repression of miRNA expression observed in human cancer (Lu et al., 2005; Ozen et al., 2008; Kumar et al., 2009).
Figure 2.
E1A-TD miRNA signature (C2C12-miR). (A) A series of pie charts summarize the E1A-TD miRNA signature, which consists of differentiation-associated (DA-miRNAs, n = 35) and proliferation-associated (PA-miRNAs, n = 21) miRNAs. Each class is subsequently distinguished according to Rb pathway responsiveness (Rb, in orange; and YH47, in green). (B) The heat map shows the regulation of miRNAs in the E1A-TD signature. Mature miRNAs expressed from multiple genomic loci are also shown (i.e., miR-133a-1 and -133a-2) to allow direct comparison with ChIP-seq data. (C) Meta-analysis of the E1A-TD miRNA signature using ChIP-seq datasets. Rows show the normalized binding of each TF at the corresponding miRNA promoter. miRNA–TF interactions experimentally validated by direct ChIP experiments are also shown (“ChIP-Validated,” see Materials and methods for details). (D) miRNA regulation upon Myc knockdown in C2C12 myoblasts was evaluated through RT-qPCR. Two different shRNAs (PLKO-Myc-KD1 and KD2) were used. Results are normalized to myoblasts expressing an shRNA against luciferase (PLKO–shLuc). U5A was used as housekeeping gene. A group of MyoD-bound muscle-specific miRNAs (miR-1, -206, and -486) was used as a negative control. The data shown are from a single representative experiment out of two repeats.
The regulation of miRNA expression during differentiation and upon E1A expression occurs at a transcriptional level, as supported by several observations. First, those miRNAs, which are organized as genomic clusters and thus share the same promoter, also shared the same pattern of regulation by E1A (Fig. 2, B and C; and Table S4); similarly, the expression of intronic miRNAs, which are usually cotranscribed with the exons of a protein-coding gene, mostly correlated with the mRNA expression pattern of their host genes (21/30, P = 0.0011; Fig. S3 A). Finally, when we compared the expression levels of mature miRNAs and their respective precursors (pri-premiRNAs) by RT-qPCR, we observed concordant regulation, both during differentiation and upon E1A expression (Fig. S3 B), which indicates that the observed miRNA expression patterns were primarily determined by transcription, rather than by posttranscriptional levels of regulation.
We classified miRNAs according to their upstream transcriptional mechanisms, using the same approach we adopted for mRNAs, i.e., by exploiting the E1A-YH47 mutant to dissect Rb-dependent and -independent effects. Almost all DA-miRNAs were Rb-independent (32/35, P = 0.0008; Fig. 2, A and B). Conversely, the expression of most PA-miRNAs was significantly dependent on Rb (17/21, P < 0.0001; Fig. 2, A and B). This latter group of miRNAs contained members of the 17-92 cluster and the 25-93-106b clusters, which have previously been shown to be E2F targets (O’Donnell et al., 2005). Finally, the Rb-independence of DA-miRNAs and Rb-dependence of PA-miRNAs were confirmed in the MSC model (34/54 DA-miRNAs, P = 0.0277; and 19/21 PA-miRNAs, P = 0.0004; Fig. S2, A and B).
Myc and MyoD regulate the expression of DA-miRNAs
To further characterize the transcriptional regulation of miRNAs of the E1A-TD miRNA signature, we examined ChIP sequencing (ChIP-seq) databases of transcription factors (TFs) relevant to the model system that we used, including Rb (Lipinski and Jacks, 1999; Chicas et al., 2010), Myc (Perna et al., 2012), and the muscle-specific TF MyoD (Cao et al., 2010).
We clustered miRNAs of the E1A-TD signature according to the TF occupancy of their promoter regions (see Materials and methods for details) and identified five major binding groups (Fig. 2 C). Overall, DA-miRNAs were poorly bound by Rb, in agreement with their Rb-independence, but were frequently bound by MyoD (group a/b) and/or Myc (group b/c/d). Conversely, PA-miRNAs, which were mainly Rb responsive, displayed a strong binding to Rb (group c′) and Myc (group c′ and d′). Importantly, a similar binding pattern for both DA- and PA-miRNAs was found in MSC-derived myotubes (Fig. S2, C and D), and almost all the previously ChIP-validated TF–promoter interactions were captured by our approach (6/6 MyoD. P < 0.0001; 6/8 Myc, P = 0.1790; and 2/3 Rb–E2F, P = 0.0240, “ChIP-validated”; Fig. 2 C and Fig. S2 C). The most frequently bound TF was Myc, even among DA-miRNA promoters (Fig. 2 C and Fig. S2 C). In agreement with this observation, the knockdown of Myc in proliferating C2C12 (where Myc is highly expressed) caused significant up-regulation of several Myc-bound DA-miRNAs (Fig. 2 D), which suggests that Myc could be largely responsible for DA-miRNA repression.
DA-miRNAs repress myoblast proliferation and promote differentiation by antagonizing the Rb–E2F pathway
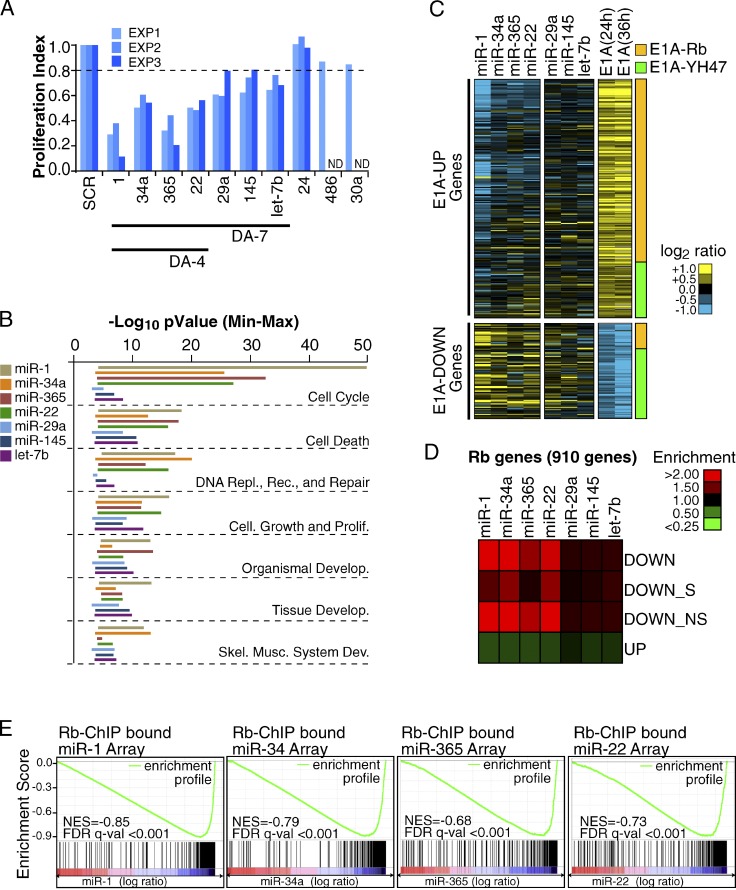
We examined whether miRNAs play a role in the establishment and maintenance of the postmitotic state. We reasoned that DA-miRNAs, the main component of our miRNA signature, might act as negative regulators of proliferation. We tested this hypothesis by overexpressing, in proliferating myoblasts, 10 DA-miRNAs belonging to the most highly expressed miRNA families found in C2C12 myotubes. Of these, seven miRNAs, hereafter referred to as “DA-7,” caused a reproducible reduction in BrdU incorporation (>20% in three independent experiments, P < 0.05), four being particularly strong in this instance (miR-1, miR-365, miR-22, and miR-34, referred to as “DA-4,” P < 0.01; Fig. 3 A).
Figure 3.
Identification of biologically relevant DA-miRNAs and characterization of their effect on gene expression. (A) Different DA-miRNAs were transfected (50 nM) in proliferating myoblasts (C2C12), and the effect on proliferation was evaluated. The bar graph shows the ratio of BrdU incorporating cells normalized over control (SCR, scrambled oligo) in three independent experiments. (B) Functional analysis of the DA-7 miRNA–regulated genes. Bars represent the Min-Max p-value (−log10 scale) of each DA-7 gene set for the most significant (P < 0.001) functional classes. (C) Gene expression changes of the 1,462 E1A genes (as from Fig. 1 B) upon DA-7 miRNA overexpression in C2C12 myoblasts. (D) The heat map shows enrichment/depletion of Rb genes among miRNA down-regulated or up-regulated transcripts. Down-regulated genes are further stratified according to the presence (DOWN_S) or absence (DOWN_NS) of seed matches in their 3′ UTR region. (E) GSEA was used to correlate Rb ChIP-bound genes (Chicas et al., 2010) with the gene expression changes observed upon miRNA overexpression. The enrichment profile (in green) together with the score (NES) and significance (FDR q-val) for each gene set analyzed are shown.
We used gene expression profiling to characterize the transcripts regulated upon overexpression of each DA-7 miRNA. In agreement with previous studies that used a similar approach (Baek et al., 2008; Selbach et al., 2008; Guo et al., 2010), each miRNA showed a widespread but mild effect on gene expression, with thousands of regulated genes being down- and up-regulated (Table 1). Although each miRNA affected different gene sets, functional classification revealed a parallel effect for all DA-7 miRNAs on proliferation and differentiation pathways (Fig. 3 B and Table S2). Indeed, by focusing on the E1A signature (as defined in Fig. 1), we observed that a significant number of E1A-induced genes were down-regulated upon DA-7 miRNA overexpression (P < 0.001 for the DA-4 subset and P < 0.01 for miR-29a, miR-145, and let-7b, which compose the remainder of DA-7 miRNAs; Fig. 3 C), which suggests that DA-7 miRNAs may cause gene expression changes opposite to those induced by the E1A oncogene.
Table 1.
DA-7 miRNA-regulated genes and miR targets
| miRNA | Down-regulated genes | Up-regulated genes | miR targets | ||||
| n | Mean ± SD | n | Mean ± SD | n | Cons. | Not Cons. | |
| miR-1 | 2,922 | −0.49 ± 0.29 | 3,326 | 0.46 ± 0.28 | 476 | 184 | 292 |
| miR-34a | 2,063 | −0.39 ± 0.20 | 2,136 | 0.40 ± 0.21 | 407 | 108 | 299 |
| miR-365 | 2,569 | −0.46 ± 0.27 | 3,142 | 0.40 ± 0.21 | 384 | 66 | 318 |
| miR-22 | 2,136 | −0.37 ± 0.18 | 2,827 | 0.41 ± 0.22 | 338 | 75 | 263 |
| miR-29a | 1,725 | −0.40 ± 0.20 | 2,188 | 0.37 ± 0.18 | 413 | 234 | 179 |
| miR-145 | 1,430 | −0.39 ± 0.21 | 2,064 | 0.39 ± 0.30 | 293 | 83 | 210 |
| let-7b | 1,991 | −0.39 ± 0.19 | 2,022 | 0.40 ± 0.22 | 355 | 199 | 156 |
The effect of DA-7 miRNAs on gene expression in proliferating myoblasts (C2C12) in three independent experiments (see Materials and methods for details) is summarized. The number (n) of genes classified as miR targets among all the genes down-regulated by each miRNA, classified as “Conserved” (Cons.) or “Not Conserved” (Not Cons.) according to TargetScan 5.1 are also shown.
Next, we concentrated on Rb genes (910 genes, as defined in Fig. 1), the major transcriptional component of the E1A signature. Rb genes were enriched within the group of transcripts down-regulated by DA-4 miRNAs and depleted within transcripts up-regulated by DA-4 miRNAs (Fig. 3 D). For the remaining DA-7 miRNAs (miR-29a, miR-145, and let-7b), we observed only a minor effect. Furthermore, the enrichment was independent of the presence of predicted miRNA–mRNA interactions within the 3′ untranslated region (UTR) or the coding sequence (CDS; Fig. 3 D and Table S5), which suggests that DA-4 miRNAs mainly acted through inhibition of the Rb pathway, rather than directly on Rb gene transcripts. Indeed, genes repressed by DA-4 miRNAs showed a significant enrichment for E2F-family binding sites in their corresponding promoter regions (P < 0.01, Table S3) and correlated remarkably well with Rb ChIP-bound genes (false discovery rate [FDR] q-value < 0.001; Fig. 3 E).
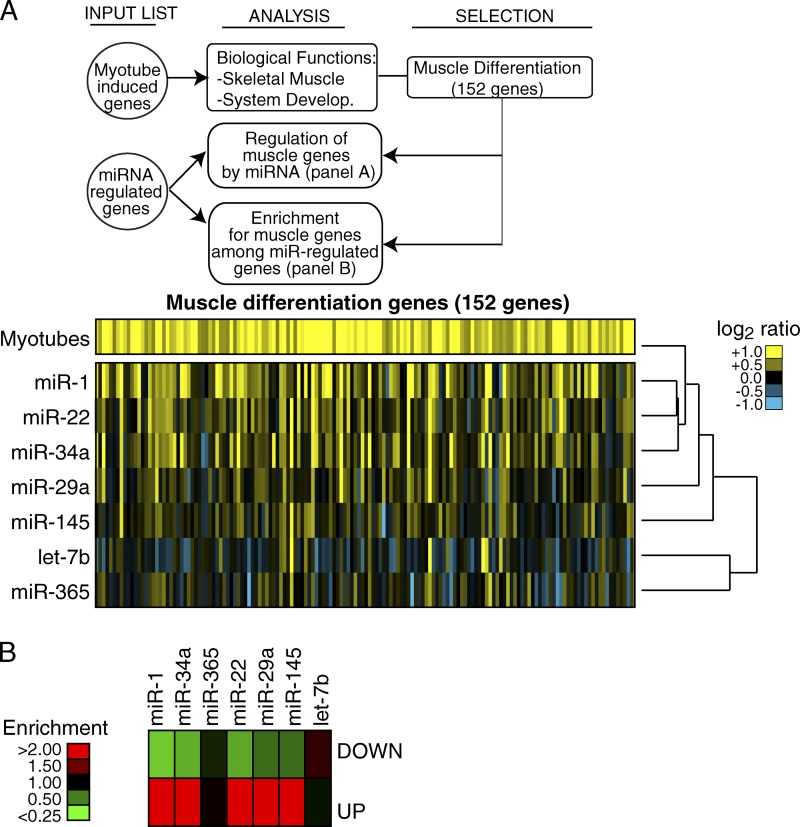
We hypothesized that DA-7 miRNA expression could also promote muscle differentiation. Indeed, many muscle genes were induced upon the expression of DA-7 miRNAs (Fig. 4), in particular by miR-1, miR-34, miR-22, and to a lesser extent by miR-29 and miR-145. Hence, muscle differentiation genes were enriched in the transcript set up-regulated by five of the DA-7 miRNAs (miR-1, -22, -34, -29, and -145, but not miR-365 and let-7; Fig. 4 B).
Figure 4.
Modulation of muscle differentiation genes by DA-7 miRNAs. A scheme summarizes the steps that were followed to analyze the regulation of muscle differentiation genes after the expression of DA-7 in proliferating C2C12 myoblasts. (A) One-way hierarchical cluster was generated by Cluster 3.0. (B) The heat map shows enrichment/depletion of muscle differentiation genes among miRNA down- or up-regulated transcripts.
DA-7 miRNAs jointly repress proliferation and accelerate the onset of muscle differentiation
In silico analyses have recently predicted that modules of coregulated miRNAs might function coordinately to ensue into a specific biological function within the cell (Tsang et al., 2010). We therefore investigated whether the miRNAs of the DA-7 module might cooperate in the regulation of proliferation and differentiation.
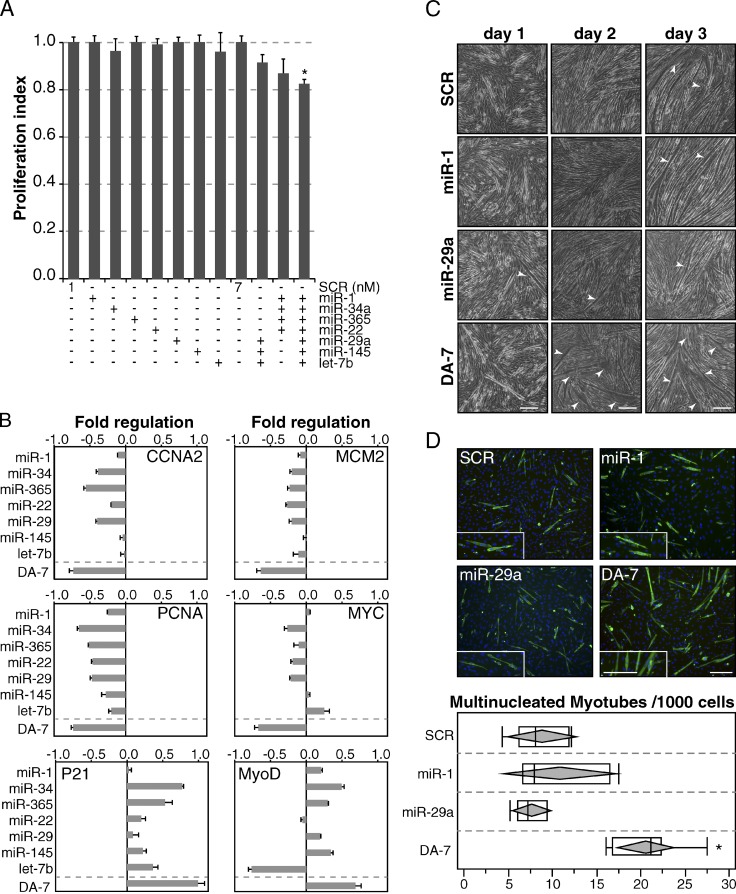
A dose-response experiment performed under conditions of increasing expression levels of individual DA-7 miRNAs revealed that very low concentrations of individual miRNAs (1 nM) had no significant effect on DNA replication (Fig. S4, A and B). At the same time, however, we noted that combinations of different miRNAs (at a 1-nM concentration) were able to repress cell proliferation, with the DA-7 subset showing a significant effect (P < 0.05; Fig. 5 A).
Figure 5.
DA-7 miRNAs cooperatively inhibit DNA replication and accelerate the onset of terminal differentiation. (A) DA-7 miRNAs were transfected at low doses (1 nM) alone or in combination in cycling C2C12 myoblasts, and the effect on proliferation was evaluated using the Delfia cell proliferation kit. Bars show the mean and standard deviation over control (SCR, scrambled oligo) in two independent experiments. The asterisk marks a significant (P < 0.05) p-value by Student’s t test. (B) The effect of DA-7 overexpression (1 nM) was analyzed by RT-qPCR (24 h after transfection) on several proliferation and differentiation genes. (C) Myotube (C2C12) formation was assessed upon transfection of DA-7 miRNAs at low doses (1 nM) in a 3-d differentiation assay. Differentiation was followed by visual inspection by phase-contrast microscopy. Enlarged/hypertrophic muscle fibers are indicated with arrowheads. (D) At day 1, multinucleated myotubes (>2 nuclei) were counted after immunostaining with MyHC antibody. Insets show a 1.5× magnification. Bars: (C) 20 µm; (D) 100 µm.
The simultaneous expression of DA-7 miRNAs repressed several proliferation genes and concomitantly induced differentiation markers either at the RNA or protein level (Fig. 5 B and Fig. S4, D and E). Of note, expression changes induced by DA-7 were stronger than those observed under conditions of overexpression of individual DA-7 miRNAs (Fig. 5 B).
Furthermore, in a differentiation assay, we observed an accelerated onset of cell differentiation into multinucleated myotubes when cells expressed the full panel of DA-7 miRNAs at low doses (1 nM). We did not observe this effect in cells overexpressing individual miRNAs related to muscle differentiation (miR-1, Chen et al., 2006; and miR-29a, Wang et al., 2008; Fig. 5, C and D).
These data suggest that, taken as a single unit, the DA-7 miRNAs are able to induce cell cycle arrest and promote differentiation.
DA-7 miRNAs directly and coordinately target genes involved in the “G1/S checkpoint” and DNA replication
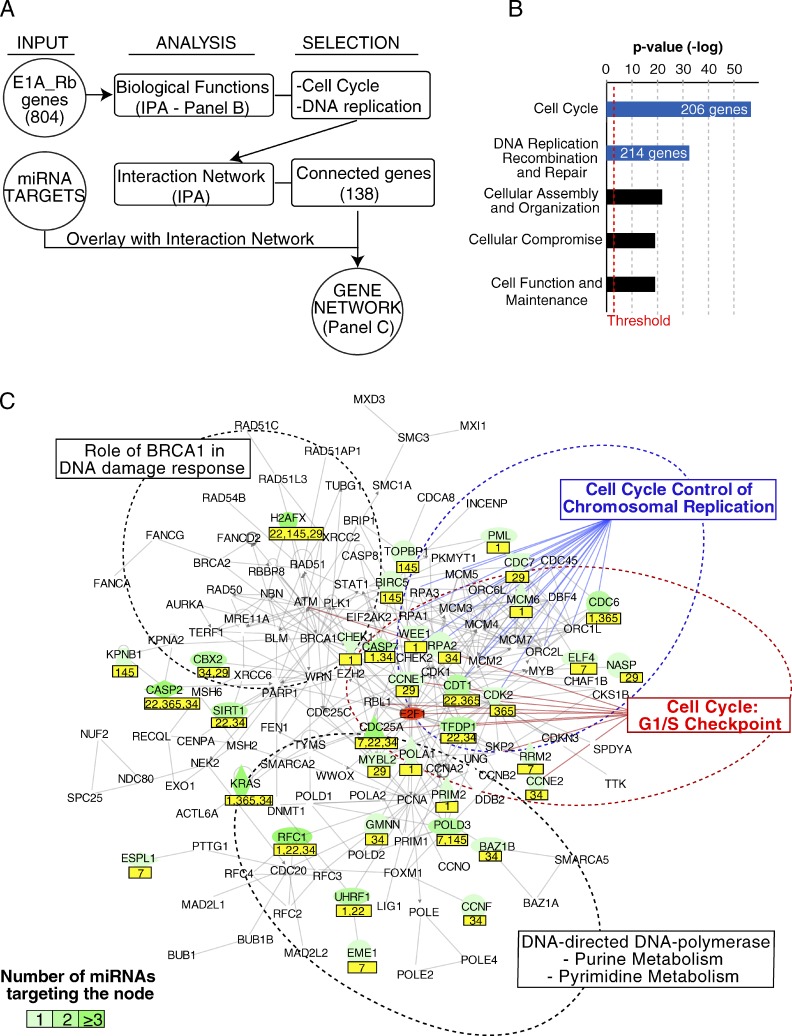
We aimed to clarify the network of miRNA–mRNA interactions through which DA-7 miRNAs (and in particular DA-4) antagonize proliferation and the Rb–E2F pathway. We developed a strategy (shown in Fig. 6) to identify direct targets of DA-7 miRNAs from our gene expression profiling, based on the evidence that mammalian miRNAs act predominantly to decrease target mRNA levels (Guo et al., 2010). In brief, we selected as candidate targets the mRNAs that (a) contain in their 3′ UTR at least one miRNA-responsive element (MRE; 8mer, 7mer-1A, or 7mer-m8), (b) were robustly expressed in the context analyzed (C2C12), and (c) were down-regulated upon forced miRNA expression in myoblasts. We isolated >300 targets (miR targets) for each miRNA (Table 1 and listed in Table S6), including several already validated by other studies/approaches (Table S6; Sethupathy et al., 2006; Xiao et al., 2009). Among miR targets, we found that a significant number of E1A-induced genes and Rb genes were targeted by miR-1 (P < 0.05), miR-22 (P < 0.001), and miR-34a (P < 0.001). Furthermore, some genes upstream of the Rb–E2F pathway could be also targeted, frequently by two or more DA-7 miRNAs (i.e., cyclinDs, CDK6, or E2Fs; Table S6). This finding suggests that, besides individual effects, DA-7 miRNAs might coordinate the coregulation of functionally connected genes to control cell cycle and DNA replication (the so-called “co-targeting” effect; Tsang et al., 2010). Therefore, we searched for pathways that were relevant for proliferation, within which we could also identify multiple mRNA–miRNA interactions. We used the Ingenuity systems pathway analysis software to identify significantly enriched biological functions within the Rb gene set (shown in Fig. 7). The two most enriched functional categories were “Cell Cycle” (206 genes, P = 2.60 × 10−57) and “DNA replication, recombination, and repair” (214, P = 3.65 × 10−33). We combined these two gene sets (n = 270) to build a network consisting of 138 proteins (nodes) and their interactions (edges; Fig. 7 C). A significant number of nodes (35 out of 138, 27.5%; P < 0.0001 as determined by a Fisher’s exact test) were miR targets (Table 1 and Table S6), and roughly one third of the nodes were targeted by at least two different DA-7 miRNAs (Fig. 7 C).
Figure 6.
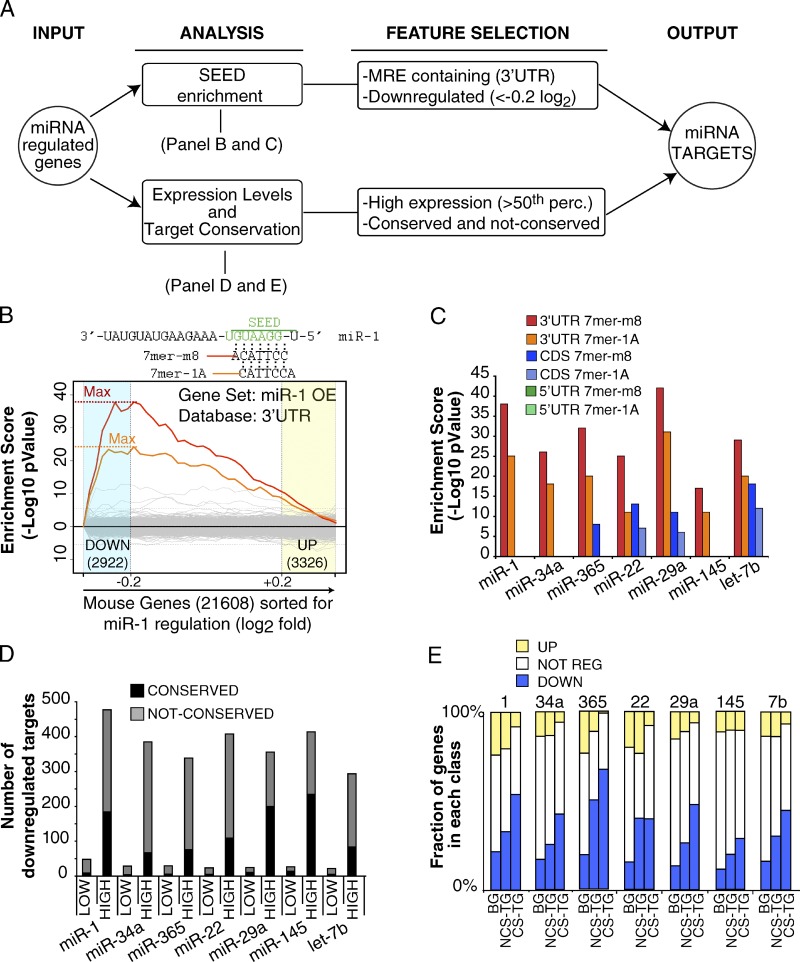
Identification of miRNA direct targets (“miR-targets”). (A) The scheme summarizes the steps that were followed to identify direct targets for DA-7 miRNAs (referred as “miR targets,” see Table S6 for a complete list). (B) Seed enrichment analysis at 3′ UTR sequences of miR-1 expression profile by Sylamer (van Dongen et al., 2008). The input list consisted of all the genes of the array (21,608), ordered from the most down-regulated to the most up-regulated upon miRNA expression. The two most overrepresented words correspond to the canonical miR-1 7mers (7mer-1A and 7mer-m8, as indicated at the top of the panel). (C) Seed enrichment analysis was repeated for each miRNA dataset with 3′ UTR, CDS, or 5′ UTR sequences. The maximal enrichment score (−log10 P-value) observed for heptamers of each overexpressed miRNA was used for comparisons and shown in the bar graph. (D) Genes containing at least one MRE (7mer words) were downloaded from the Human TargetScan database (Release 5.1; mouse predictions) and split according to the expression level in myoblasts (LOW/HIGH based on the 50th percentile of the average raw signal). The bar graph shows the number of MRE-containing genes (conserved or not conserved, according to TargetScan) that were down-regulated upon each DA-7 miRNA overexpression. (E) Robustly expressed genes (HIGH, as for D) were stratified according to the presence of conserved MRE (CS-TG) or nonconserved MRE (NCS-TG). The histogram shows the distribution of up-regulated (UP), down-regulated (DOWN), and not regulated (NOT REG) genes for each class. The overall distribution of all genes (Background, BG) is also shown as a reference. A significant response (P < 0.05, Fisher’s exact test) was observed for genes with either conserved or nonconserved sites.
Figure 7.
Network of miR targets. (A) The scheme summarizes the steps that were followed to generate a network of DA-7–targeted genes involved in the control of proliferation and DNA replication. (B) The bar graph shows the most enriched biological functions as assessed through Ingenuity systems pathway analysis software. The list of E1A_Rb–induced genes (804) was used as input. A red line marks the threshold for significance. (C) The gene network of 138 proteins (from the two most enriched biological functions as determined in B) and their connections (gray lines) is depicted. On top of this map, miR targets are displayed in green. miRNA names are reported in a yellow box below the node. A dotted line marks areas with nodes belonging to canonical pathways.
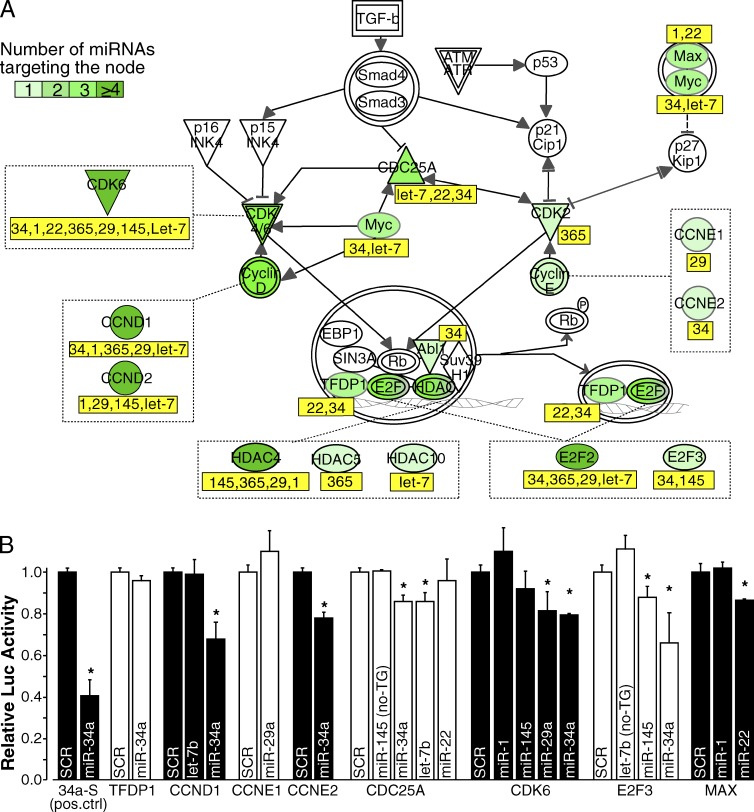
Within this network, we focused on two specific canonical pathways relevant to the control of proliferation: the G1/S checkpoint (nine nodes, five of which were targeted by DA-7 miRNAs) and “Cell cycle control of chromosomal replication” (19 nodes, of which 6 were DA-7 miRNA targets). A canonical representation of these two pathways shows that both “activators” of the G1/S checkpoint (positive regulators of the pathway, such as cyclins and CDKs; Fig. 8 A) and structural or catalytic components of the DNA replication machinery (MCMs, RPA, Pol A, and Pol D; unpublished data) were targeted by DA-7 miRNAs, which suggests a synergistic effect of the entire DA-7 class in silencing these two pathways. To measure this effect quantitatively, we compared the mean number of miRNA–mRNA interactions of the DA-7 subset of miRNAs with that of other miRNAs. We observed a significant enrichment of either target genes (a mean of 5.43 targets per miRNA; P = 0.0013) or MREs (an average of 8.14 MREs per miRNA, P = 0.0014) within the G1/S checkpoint pathway (Fig. S5, A and B), but not within the “Cell cycle control of chromosomal replication” (targets, P = 0.4095; MRE, P = 0.4035). Many of these miRNA–mRNA interactions are predicted to be conserved in higher eukaryotes (a mean of 4.90 conserved targets per miRNA).
Figure 8.
DA-7 miRNAs target G1/S checkpoint genes. (A) Representation of the G1/S checkpoint canonical pathway. Genes targeted by at least one of the DA-7 miRNA are highlighted in green with a color code (from light to dark green) according to the number of miRNA–mRNA interactions. Those DA-7 miRNAs targeting each node are reported below in the yellow box. (B) Relative firefly luciferase activity of 3′ UTR reporter constructs for some genes of the G1/S checkpoint, after transfection with selected DA-7 synthetic miRNA mimics. Error bars represent standard deviations from three independent transfections. Asterisks mark significant values (P < 0.05; Student’s t test). Two predicted interactions (CDC25A–miR-145 and E2F3–let-7b), for which we did not observed a decrease in mRNA levels upon DA-7 expression (and, therefore, were not considered as miR targets), were also analyzed (no-TG).
We generated reporter plasmids for several miRNAs by cloning portions of the 3′ UTRs encompassing their predicted MREs downstream of a luciferase reporter gene; this tool allowed us to assess miRNA–mRNA interactions directly by measuring relative luciferase activity. In 9 out of 16 predicted interactions (56%), luciferase activity of the reporter was indeed significantly reduced by transfection of the corresponding DA-7 miRNAs into HEK293T cells (Fig. 8 B and Table S7).
DA-7 miRNAs inhibit E1A-induced transcriptional reprogramming and the induction of cell cycle reentry
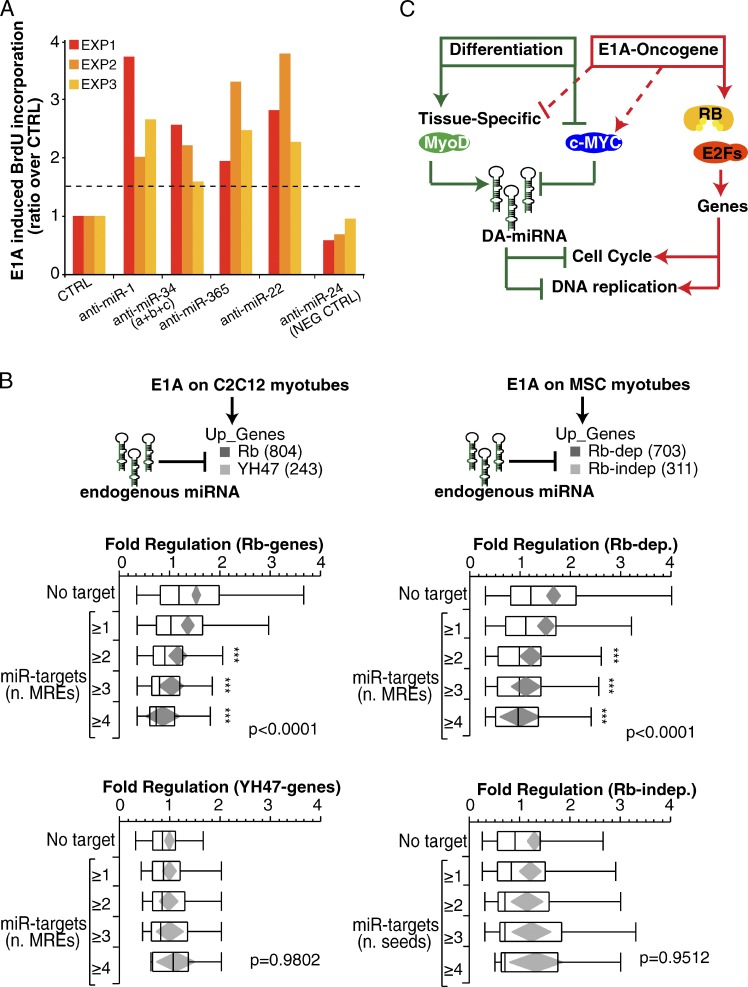
Because DA-7 miRNAs and the E1A oncogene had opposing effects on the control of proliferation, we reasoned that the ablation of DA-7 miRNAs in myotubes should enhance cell cycle reentry by E1A. We tested this hypothesis directly, by knocking down the strongest DA-7 miRNAs (i.e., the DA-4 miRNAs: miR-1, miR-34, miR-365, and miR-22) with anti-miRs and measuring cell cycle reentry upon E1A expression in differentiated cells. For all tested miRNAs, we scored a significant and reproducible (P < 0.05 by Student’s t test; n = 3) increase in BrdU-positive cells (Fig. 9 A). Conversely, no effect was observed by knocking down miR-24, which also scored negatively in overexpression experiments (Fig. 3 A and Fig. 9 A). Therefore, DA-4 miRNAs at their physiological levels are able to restrict unscheduled cell proliferation induced by the E1A oncogene.
Figure 9.
Endogenous miRNAs inhibit oncogene-induced reprogramming. (A) miRNA inhibitors (anti-miRs) were transfected (100 nM) in differentiated myotubes (MSC) infected with suboptimal doses (MOI: 50) of Ad-E1A virus. The effect on proliferation was evaluated by means of BrdU incorporation. The bar graph shows the ratio of the BrdU-incorporating cell normalized over control (SCR, scrambled oligo) in three independent experiments. Anti–miR-24 was used as negative control. (B) The effect of endogenous DA-7 on E1A-induced genes was evaluated in C2C12 (left) and MSC (right) myotubes. E1A-induced genes were distinguished into two transcriptional classes (Rb and YH47 genes, for C2C12; and Rb-dependent and Rb-independent genes, for MSC), and each gene set was further stratified according to the miR target list (Tables 1 and S6) and the number of MREs present in each 3′ UTR region. Also reported are the mean diamonds (in gray) and the p-value (ANOVA). Three asterisks mark significant distribution (P < 0.05) according to Dunnet’s t test. (C) A scheme summarizes the role of DA-7 miRNAs to antagonize reactivation of Rb genes involved in cell cycle and DNA replication.
DA-7 and E1A also had opposing effects on gene expression, and in particular on the Rb-dependent component (Fig. 3 C); thus we analyzed whether endogenous DA-7 miRNAs could repress their Rb target genes during cell cycle reentry upon E1A expression in differentiated cells. At 24 h after infection, when we evaluated the gene expression by E1A, many DA-7 miRNAs were still expressed at higher levels compared with cycling myoblasts (Fig. S5 C), which indicates that they might still be active on their targets. We therefore stratified E1A genes by the number of MREs for DA-7 miRNAs and observed that mRNAs induced by E1A were significantly less induced if they interacted with DA-7 miRNAs (P < 0.0001), with a dose-dependent effect (the higher the number of MREs, the lower the induction of genes; Fig. 9 B, “E1A on C2C12 myotubes”). This effect was not dependent on the length of the 3′ UTR regions (adjusted P < 0.001) and was specifically restricted to Rb genes, as YH47 genes were all equally regulated by E1A regardless of the presence and the number of MREs (Fig. 9 B). Importantly, we were also able to reproduce these findings in primary myotubes: upon E1A expression, the induction of Rb-dependent genes with MREs responsive to DA-7 miRNAs was dampened down in a seed-dependent fashion, whereas no significant effect was observed on Rb-independent genes (Fig. 9 B, “E1A on MSC myotubes”).
These results show that DA-7 miRNAs play a role in preventing cell-cycle reentry by curbing changes in gene expression that are induced by the E1A oncogene and confirm that there exists an antagonism between DA-7 miRNAs and the Rb pathway, even when cells express endogenous levels of miRNAs (Fig. 9 C).
Discussion
A coordinated mRNA/miRNA expression program in the control of proliferation and differentiation
We have uncovered a coordinated mechanism of control of proliferation and differentiation, which couples two distinct processes: the transcriptional regulation of cell cycle and DNA replication genes, which lies under the control of the Rb–E2F pathway, and an miRNA pathway associated with differentiation (DA-miRNAs), which targets a significant fraction of the same genes, thus conferring robustness to the phenotypic output.
Our results confirm that the key effect of E1A at the transcriptional level is its interference with Rb and its cognate proteins (p107 and p130, “pocket-proteins), which are involved in both differentiation and cell cycle withdrawal (Lipinski and Jacks, 1999; Ferrari et al., 2008). In parallel, the E1A-associated miRNA expression program consists of a widespread repression of DA-miRNAs in an Rb-independent fashion.
During muscle differentiation, the induction of many tissue-specific miRNAs (so-called “myomirs”) lies under the control of muscle-specific regulatory factors (MRFs), such as MyoD and myogenin (Williams et al., 2009). We observed an Rb-independent regulation of myomirs (miR-1, miR-133, miR-206, and miR-486), which is in agreement with the known ability of E1A to interfere with muscle-specific transcription through its N-terminal region, independently of its interaction with pocket proteins (Tiainen et al., 1996; Puri et al., 1997; Nicassio et al., 2005).
Our work also demonstrates Myc-dependent repression of several DA-miRNAs. Myc is down-regulated during muscle differentiation (Yeilding et al., 1998), whereas it is rapidly induced when cells reenter a proliferative state from quiescence or terminal differentiation (for reviews see Amati et al., 1998; Grandori et al., 2000). We also observed that many Myc-dependent miRNAs are induced in serum-starved primary fibroblasts, which are quiescent, compared with proliferating cells (unpublished data). Consistently, Myc overexpression in other cellular systems has been shown to repress the transcription of the same miRNAs (Chang et al., 2008). Of note, Myc itself is a direct target of two miRNAs (miR-34 and miR-145) that are under the transcriptional control of p53 (He et al., 2007; Sachdeva et al., 2009; Cannell et al., 2010), which is required for complete muscle differentiation (Porrello et al., 2000). Hence, it is tempting to speculate that the coordinated regulation of miRNAs by Myc and p53 might be generally involved in the control of cell cycle exit/reentry in nonproliferative contexts (i.e., differentiation, quiescence, and senescence) as well as in cancer (Chang et al., 2008; Kota et al., 2009).
A module of differentiation associated miRNAs act as “quiescence keepers”
We demonstrated that several DA-miRNAs (the DA-7 miRNAs) play a role in the control of proliferation. When overexpressed in proliferating myoblasts, where they are normally expressed at low levels, these DA-7 miRNAs induce cell-cycle arrest; instead, the inhibition of the expression of four of the seven DA-7 miRNAs in myotubes, where they are normally abundantly expressed, is sufficient to enhance E1A-induced DNA replication. These miRNAs reinforce the postmitotic barrier against oncogene-induced proliferation. Therefore, we propose the term “quiescence keepers” to describe these miRNAs. Besides individual effects, the quiescence-keeper miRNAs also cooperate to induce cell cycle arrest and differentiation. The combination of DA-7 had a significant biological effect even when miRNAs were expressed at very low (paraphysiological) levels, which suggests that the joint regulation of different miRNAs has stronger activity than individual miRNAs, as observed in previous studies (Marasa et al., 2009).
We identified a set of putative direct gene targets (“miR targets”) for each quiescence-keeper miRNA, and we built a network of miRNA–mRNA interactions that reveals how miRNA and gene expression pathways intersect to regulate cell cycle exit/reentry. We found that miRNAs target several key nodes of the Rb–E2F network, including cyclins and Cdks, which regulate cell cycle checkpoints and Rb phosphorylation (Sherr, 1996), as well as E2Fs and DP1 TFs, which mediate the transcription of genes necessary for cell cycle progression (Trimarchi and Lees, 2002). In addition, downstream effectors of the Rb pathway are also targeted by miRNAs, thus revealing multiple levels of interaction between miRNAs and Rb-dependent genes that contribute to maintain the balance of the postmitotic state.
In silico analyses have recently predicted that miRNAs might function coordinately within a given pathway by cotargeting several genes involved in the same biological function (Tsang et al., 2010). We have now demonstrated this same concept experimentally, showing that functionally related miRNAs (the quiescence keepers) act by cotargeting genes of the G1/S transition to reinforce the inhibition of cell cycle progression and accelerate the onset of differentiation.
We believe that our approach, which consists of networking a “group of co-regulated miRNAs” (with a similar phenotype) to a “group of co-targeted genes” (involved in the phenotype), can be applied to other experimental settings to reveal functional mechanisms that would be difficult to uncover with a “classical” single gene/miRNA approach.
From differentiation to tumors: could quiescence keepers be tumor-suppressor miRNAs?
Several studies suggest that a reduction in miRNA levels is functional to cell transformation, although the phenotypic advantages afforded to cells by reduced miRNA expression remain unclear (Kumar et al., 2009; Martello et al., 2010). Our results suggest that one such advantage may be the loss of control of quiescence. In fact, by inhibiting the reexpression of oncogene-induced genes in a seed-dependent fashion, quiescence-keeper miRNAs work as a safety net against transformation. We have shown how a module of endogenous miRNAs can counteract the transcriptional alterations induced by an oncogene (E1A, in this case), and we suggest that the miRNA pathway may not only serve to stabilize the transcriptional output of a cell (“buffering” effect), but may also work in preventing undesirable alterations because of oncogenic disruption of normal cell function (“keeper” effect).
We speculate that other tissue/context-specific quiescence-keeper miRNAs exist, which are likely to contribute to tumor suppression through the regulation of proliferation and differentiation. Their identification would provide a spectrum of potentially valuable tumor markers. In this regard, we note that tumors with low levels of expression of miRNAs tend to be associated with a poorer prognosis than those with normal miRNA levels (Lu et al., 2005; Merritt et al., 2008), which suggests that patient stratification on the basis of expression levels of quiescence-keeper miRNAs may be helpful for prognostic purposes.
A deeper understanding of the mechanisms responsible for determining the balance between differentiation and cell proliferation may help to develop new therapeutic strategies in cancer. Indeed, the reexpression of quiescence keepers in tumors can induce cell-cycle arrest and differentiation of cancer cells (Kumar et al., 2008; Kota et al., 2009; Taulli et al., 2009; Ting et al., 2010), and animal tumor models have been recently used to reexpress tumor-suppressive quiescence-keeper miRNAs (miR-34, let-7, and miR-1) with promising results (Kim et al., 2011; Liu et al., 2011; Trang et al., 2011; Wu, 2011). Thus, we envision that quiescence keepers might hold a key to new strategies for differentiation therapy in cancer.
Materials and methods
Cell culture procedures
C2C12 and MSC-RbloxP/loxP myoblasts were differentiated in vitro to myotubes as described previously (Camarda et al., 2004; Nicassio et al., 2005). In brief, mouse MSCs were induced to differentiate by plating cells (150,000/35 mm) on gelatin-coated dishes (Iwaki), cultured in DME supplemented with 10% FBS, and, 24 h later, switched to 5% horse serum for 48 h. Mouse C2C12 myoblasts were induced to differentiate by plating the cells (350,000/35 mm) in collagen-coated dishes (Iwaki) and culturing them in DME supplemented with 2% horse serum for 3 d. To eliminate undifferentiated cells, 50 mM cytosine-d-arabinofuranoside was added for the first 48 h.
Myotubes were infected with an adenovirus carrying the 12S form of E1A (dl520 amino acid changes: del 140–186; named E1A), with the YH47/dl928 E1A mutant (YH47/dl928, amino acid changes: Y47H and C124G; named YH47) or with a control adenovirus expressing a deletion of essentially the entire E1A gene (dl312 amino acid changes: del 1–289; named MOCK).
Multiplicity of infection (MOI) was 300 for all the viruses. The depletion of the floxed Rb gene in MSC Rbflox/flox myotubes (provided by A. Berns and M. Vooijs, The Netherlands Cancer Institute, Amsterdam, Netherlands) was achieved by Ad-CRE infection at MOI 1,000 (Camarda et al., 2004). Correct differentiation of myotubes was verified by immunophenotypic analysis using an antibody against a muscle-specific protein (myosin heavy chain [MyHC]) and by RT-qPCR of early and late differentiation markers (MyoD and MyoHC genes, respectively). Induction of cell cycle reentry by E1A was assessed by continuous measurement of BrdU (20 µM) incorporation over 24 h. MyHC-positive cells were counted to ensure that only fully differentiated cells were considered. Antibodies used in immunofluorescence: BrdU, clone Bu20a (DakoCytomation); and MyHC, rabbit polyclonal or mouse monoclonal (MF-20; a gift from G. Cossu, University College London, London, England, UK). Secondary antibodies were purchased from Molecular Probes or Invitrogen. Nuclei were counterstained with Hoechst 33258 or DAPI. Antibodies used in Western blotting: pRb, clone G3-245 (BD); β-tubulin, clone 2-28-33 (Sigma-Aldrich); Myc, clone Y69 (catalogue no. ab32072; Abcam); PCNA (catalogue no. 610664; BD); MyoD, C-20 (catalogue no. SC-304; Santa Cruz Biotechnology, Inc.); Myogenin, F5D (catalogue no. SC-12732; Santa Cruz Biotechnology, Inc.); NP95 (home-made rabbit polyclonal); and Vinculin (catalogue no. V9131; Sigma-Aldrich). Detection was performed with HRP-conjugated secondary antibodies (Cappel or Bio-Rad Laboratories) and ECL Western blotting reagent (GE Healthcare) or a West Dura kit (Thermo Fisher Scientific). Chemiluminescence of Vinculin, Myc, PCNA, MyoD, NP95, and Myog was performed with the ChemiDoc XRS+ (Bio-Rad Laboratories), and band densitometry was analyzed with Image Laboratory 3.0.1 (Bio-Rad Laboratories).
Microscopy
Visual inspection of differentiation was performed under a light microscope (DM5500B; Leica) using a 5× objective lens (NA 0.15) at RT and using Mowiol mounting medium. BrdU and MyHC staining were viewed under a fluorescent microscope (DM5500B; Leica) using a 5×/NA 0.15 objective lens at RT. Fluorochromes used were Cy3 (for BrdU) and Alexa Fluor 488 (for MyHC). All images were acquired with a camera (DFC350FX; Leica) and LAS-AF image software (Leica). Images were processed with ImageJ, and imaged figures were constructed in Illustrator (Adobe).
miRNA transfection
For experiments shown in Fig. 3 and Fig. 4, MiScript miRNA mimics (QIAGEN) were transfected at 50 nM into proliferating C2C12 cells, with HiPerFect reagent (QIAGEN). After 36 h, RNA was collected for gene expression analysis and a BrdU (20 µM) incorporation assay (1 h, 30 min pulse) was performed. For experiments shown in Fig. 9, miRNA knockdown in MSC-derived myotubes was obtained by transfection of 100 nM miRIDIAN miRNA inhibitors (Thermo Fisher Scientific). 1 h before transfection, myotubes were infected with the dl520 adenovirus at MOI 50. Cell cycle reentry was evaluated by measurement of BrdU (20 µM) incorporation at 40 h after infection.
For experiments shown in Fig. 5, C2C12 cells (p28) were transfected with control (scrambled) or 1 nM miRNA mimics. For the differentiation assay, cells were plated on gelatin-coated dishes (400,000 cells/35 mm) and switched (day 1, 24 h after transfection) to differentiation medium (2% horse serum). Multinucleated (>2 nuclei) myotubes were assessed either by visual inspection using phase-contrast microscopy or by immunostaining with MyHC antibody (MF20) and DAPI.
Assessment of BrdU incorporation using the Delfia cell proliferation kit
Proliferating C2C12 cells (p28) were plated in 96-well plates (Viewplate, catalogue no. 6005181; PerkinElmer) at a concentration of 4,000 cells/well. Cell were transfected in triplicate with miScript miRNA mimics (QIAGEN) at a final concentration ranging from 0.5 nM to 50 nM in a final volume of 200 µl with HiPerFect reagent (QIAGEN) and using the reverse transfection protocol according to manufacturer’s instructions. When assessing the combinatorial effect of DA-7 miRNAs, mimics were diluted together to give a final concentration of 1 nM each. At 34 h after transfection, BrdU was added to the medium (2 h pulse), and incorporation was measured subsequently using the Delfia cell proliferation kit (PerkinElmer) according to the manufacturers’ instructions and using the EnVision Multilabel Reader (PerkinElmer).
Luciferase reporter assays
HEK293T cells (280,000) were plated in a 24-well plate. Shortly after plating, cells were transfected with 150 ng of the indicated 3′ UTR reporter construct (pmiR-GLO; Promega) and miRNA mimics (QIAGEN) at a 50-nM final concentration using Lipofectamine 2000 (1.5 µl/well; Invitrogen) according to the manufacturer’s instructions. After 24 h, cells were lysed and assayed in triplicate wells for firefly and renilla luciferase activity using the Dual-Luciferase Reporter Assay System (Promega) and an EnVision Multilabel plate reader. Firefly luciferase activity was normalized to renilla luciferase activity for each transfected well as follows: [firefly(miR)/renilla(miR)]/[firefly(SCR)/renilla(SCR)]. Data are representative of two or three independent experiments performed on different days (see also Table S7).
Gene expression analysis
Gene expression of E1A-infected C2C12-myotubes was performed through the Affymetrix GeneChip Mouse genome 430 2.0 platform. Raw data together with detailed description of the normalization procedure is available in the GEO database (GSE28457). Significantly regulated genes were selected in two independent biological replicates, each performed in two technical replicates. The criteria for inclusion in the list of “E1A regulated genes” were: >0.25 log2 ratio for up-regulated genes and <−0.25 log2 ratio for down-regulated genes, and P ≤ 0.05 by Welch’s t test with Benjamini and Hochberg multiple testing correction. A ratio between the regulation of the YH47 mutant and wt-E1A was used to derive two classes of regulation for E1A genes: Rb genes (YH47/E1A < 0.40) and YH47 genes (YH47/E1A ≥ 0.40). Gene expression of E1A-infected MSC-RbloxP/loxP myotubes was performed as described previously (Camarda et al., 2004). In brief, total mRNA was converted into cDNA and then biotin-labeled cRNA using the SuperScript II (Invitrogen) and ENZO (Affymetrix) kits. cRNAs were hybridized to MGU74Av2 oligonucleotide microarrays (Affymetrix) according to manufacturer’s instructions. Data analysis was performed using Affymetrix Microarray Suite v.5.0 and Excel software (Microsoft). E1A-regulated genes (>0.25 log2 ratio for up-regulated genes and <−0.25 log2 ratio for down-regulated genes, P ≤ 0.05 by Welch’s t test) were classified in two classes according to the ratio between Rb−/− MSC myotubes and E1A-infected MSC myotubes: Rb-dependent genes (Rb/E1A ≥ 0.40) and Rb-independent genes (Rb/E1A < 0.40).
Gene expression profiling upon miRNA overexpression was performed by Affymetrix Mouse Gene 1.0 ST arrays. Three biological samples were analyzed for each condition. Regulated genes were selected in matched experiments as follows: >0.2 mean log2 ratio for up-regulated genes and <−0.2 mean log2 ratio for down-regulated genes, P < 0.05 by Welch’s t test with Benjamini and Hochberg multiple testing correction. Data are available in the GEO omnibus database (GSE26764).
miRNA expression analysis
The E1A-TD miRNA signature was obtained with a TaqMan Low Density Array microRNA Signature Panel V2.0 (Applied Biosystems). Total RNA from two independent biological replicas (either for C2C12 or MSC myotubes) was used for miRNA profiling. Raw data (Ct values) were exported in Excel (Microsoft) for filtering and normalization. Low (mean Ct > 30.01) and not expressed miRNAs (i.e., “Not Amplified”) were excluded from the analysis. Then, median expression of housekeeping genes (U6b, Sno135, and Sno202) present on the array was used for data normalization. Regulated miRNAs were selected based on the following criteria: (a) DA-miRNAs, induced upon differentiation (>0.25 log2 ratio between myotubes and myoblasts) and repressed by E1A expression (<−0.25 log2 ratio between E1A vs. mock infected myotubes), and (b) PA-miRNAs, repressed upon differentiation (<−0.25 log2 ratio between E1A vs. mock infected myotubes and myoblasts) and induced by E1A expression (>0.25 log2 ratio E1A infected [E1A] vs. mock-infected myotubes). Classification of miRNAs as Rb-dependent or Rb-independent was performed as for mRNAs. Several miRNAs were analyzed as precursors (pri- and premiRNAs) or mature miRNAs by means of RT-qPCR performed with the miScript System (QIAGEN; shown in Fig. S3 B). Reactions for mature miRNAs were performed using 5 ng cDNA for each reaction and U6b/U5a as housekeeping genes. miRNA precursors were measured by using 20 ng cDNA for each reaction and 18s as housekeeping gene.
GSEA
GSEA analysis was performed using the preranked gene list (based on log2 expression ratio) of the E1A regulated genes (i.e., the 1,462 genes, Fig. 1 E) or miRNA-regulated genes (Fig. 3 E). Those genes bound by Rb both in quiescent and senescent cells according to ChIP-seq experiments (Chicas et al., 2010) were used as gene sets to calculate the enrichment score (ES). P-values were calculated by performing 1,000 random permutations of genes labels to create ES null distribution.
TF analysis (TRANSFAC)
E1A-TD mRNA signatures from C2C12 or MSC myotubes or miR-regulated genes (C2C12) were analyzed for TF binding sites, as defined in TRANSFAC version 7.4. Specifically, we used the “compute overlap” tool at the Molecular Signatures Database (MSigDB; Subramanian et al., 2005). Only significantly enriched gene sets (P < 0.01) matching known TFs were considered.
ChIP-seq meta-analysis of E1A genes
High-confidence Rb-binding regions were previously assessed through ChIP-seq in proliferating, quiescent, and senescent human IMR90 fibroblasts (Chicas et al., 2010). Binding regions were mapped to the human genome assembly (NCBI36/hg18) by associating each Ref_seq to a promoter region (considering the 1,000 bp upstream or downstream of the transcription start site). In particular, we considered those bound both in quiescent and senescent cells as Rb-bound genes. Finally, Human Entrez_IDs of these genes were converted into Mouse Entrez_IDs to classify E1A genes or miR-regulated genes as Rb “ChIP-bound” or “Not-bound.”
Genome-wide binding of MyoD, Myc, and Rb to miRNA promoters
To identify TFs associated with promoters of the E1A-TD miRNA signature, a three-step procedure was followed.
First, we retrieved MyoD-, Myc-, and Rb-binding regions, as assessed by Chip-seq experiments available from the literature: MyoD (mouse, C2C12; Cao et al., 2010), Myc (mouse, 3T9-MEF; Perna et al., 2012), and Rb (human, IMR90; Chicas et al., 2010). Only high-confidence binding regions, as determined by each original contribution, were used. Then, we retrieved miRNA transcription start sites (TSS) along with their TSS scores from the work of Marson et al. (2008). TSS scores below zero were excluded from subsequent analyses. Of note, we re-annotated the genomic coordinates of some well-characterized muscle-specific miRNA promoters (miR-1-1, miR-1-2, miR-133a-1, miR-133a-2, miR-206, and miR-486; Rao et al., 2006; Small et al., 2010), as data produced in embryonic stem cells were likely to be inaccurate in the case of these highly tissue-specific miRNAs. All the genomic coordinates were converted to the last genome assembly (NCBI Build 36.1/hg18 and NCBI Build 37/mm9) using the liftOver tool of the UCSC genome browser. Overall, 550 promoters were available for human miRNAs and 393 for mouse miRNAs. Because the genomic intervals corresponding to each TSS varied from 200 bp to several kilobases, a region of 2,000 bp centered on the middle of the TSS region was selected (“miR promoter”). Finally, genomic intervals corresponding to TF binding and to miR promoters were joined, using mouse promoters for MyoD and Myc and human promoters for Rb. Overlapping regions (at least 1bp) were considered as “TF-bound miR promoters.” When multiple peaks were found on a given promoter, only the highest was considered. The height of the highest peak was finally normalized in a 1–100 scale to allow their representation using the same scale for different ChIP-seq experiments and reported in a log2 scale (Fig. 2 C and Fig. S2 C).
ChIP validated miRNA–TF interactions
Several miRNA–TF interactions shown in Fig. 2 C and Fig. S2 C were experimentally validated by direct ChIP experiments (“ChIP validated” columns) according to the following references: MyoD (Cao et al., 2006; Rao et al., 2006; Small et al., 2010), MYC (O’Donnell et al., 2005; Chang et al., 2008; Gao et al., 2009), and E2F1 (Woods et al., 2007; Bueno et al., 2010).
MYC knockdown in proliferating C2C12
Experiments were conducted using a pLKO.1 lentiviral vector expressing two different hairpins complementary to mouse Myc or, as a control, a hairpin complementary to luciferase. These vectors were a gift of B. Amati (Istituto Europeo di Oncologia, Milan, Italy). The sequences used for MYC RNAi were: Myc KD1, 5′-CTGGAGATGATGACCGAGTTA-3′; and Myc KD2, 5′-CCTGAAGCAGATCAGCAACAA-3′. After four rounds of infection, cells were selected with 2 µg/ml puromycin. Total RNA was collected after 3 d of selection, and Myc knockdown was evaluated by RT-qPCR using a QuantiTect Primer Assay (QT00096194, QIAGEN).
Seed enrichment analysis by Sylamer
All the Ref_seq expressed in C2C12 cells from the Affymetrix Mouse Gene 1.0 ST arrays (21608) were ordered from the most down-regulated to the most up-regulated upon overexpression of each miRNA in C2C12 myoblasts. This list was uploaded in Sylamer (van Dongen et al., 2008) to compute word enrichment. The analysis was performed with sequences of the 3′ UTR, which usually contains the MREs, as well as with the CDSs or the 5′ UTRs for the mouse genome (assembly: NCBI37/mm9). Word sizes of 6, 7, and 8 were analyzed with standard parameters. The maximal enrichment score (−log10 p-value) observed for heptamers of each overexpressed miRNA was used for comparisons. For each dataset, the highest enrichment was observed in 3′ UTR sequences within the threshold used to define “down-regulated” genes (−0.2 log2 ratio), and the most represented words (7mers) exactly matched with the canonical seed (7mer-1A and 7mer-m8) of the overexpressed miRNA (Fig. 6 B). Some of the miRNA-regulated gene lists (miR-22, miR-29, and let-7b) also showed a weaker, but significant, enrichment of MREs in the CDSs (Fig. 6 C), which is consistent with previous evidence that a minor fraction of miRNA binding sites are located within the CDS (Lewis et al., 2005; Easow et al., 2007). None of the miRNAs showed enrichment in 5′ UTR sequences.
Ingenuity pathway functional analysis
Significantly enriched functional classes (Figs. 1 C, 3 B, 7 B, and S1 C) were identified through Ingenuity systems pathway analysis (Ingenuity Systems), running a core analysis, and considering the enriched functions (“Molecular and cellular functions” and “Physiological system development and function,” P < 0.001) by using as input E1A-regulated or miR-regulated genes. Each significant functional class corresponds to several significant (P < 0.001) gene ontology (GO) terms. Accordingly, for each class the minimum (MIN) and maximum (MAX) p-value of the corresponding GO terms was reported. The complete lists of all the significant functional classes for E1A-regulated or miR-regulated genes are reported in Table S2. To generate a network of cell cycle and DNA replication genes, the two most enriched functional categories of E1A Rb-dependent genes (cell cycle, 206 genes, P = 2.60 × 10−57; and DNA replication, recombination, and repair, 214 genes, P = 3.65 × 10−33; Fig. 7 B) were selected. The combined (n = 270) list was used to build a network consisting of 138 proteins (nodes) and their interactions (edges) using the “connect” tool (with standard parameters). The canonical pathways of “G1/S checkpoint” and “Cell cycle control of chromosomal replication” were selected from the network and analyzed separately for mRNA–miRNA interactions.
miRNA target enrichment analysis
The mean number of either target genes or MREs (Fig. S5, A and B) for each pathway was determined as the ratio between the total number of elements targeted (genes or MREs) within a pathway (N) and the number of miRNAs of each class (n), as follows: ALL, all the miRNA families included in the TargetScan 5.1 database (n = 373); PA, proliferation associated miRNA families (n = 13); DA, differentiation associated miRNA families (n = 26); and DA-7 miRNA families (n = 7). The following example illustrates the procedure: for all the 373 miRNA families listed in TargetScan, we found a total of 1,716 MREs (257 conserved and 1,459 nonconserved) in the “G1/S checkpoint,” and 780 MREs (42 conserved and 738 nonconserved) in the “Cell cycle control of chromosomal replication.” Thus, on average, a single miRNA targets 4.6 G1/S and 2.0 DNA replication MREs. Conversely, DA-7 miRNAs target a total of 57 G1/S and 16 DNA replication MREs, with a mean of 8.14 and 2.3, respectively. Because all DA-7 miRNAs belong to the “broadly conserved” group of miRNAs, we repeated the analysis considering only broadly conserved miRNA families, as defined in TargetScan, (n = 86). We obtained almost identical results for all the analyses performed. To determine the significance of the difference observed, we generated 10,000 random lists (R-project software) of seven miRNAs (chosen from the 373 miRNA families listed in TargetScan), and we calculated how frequently a group of seven miRNAs randomly selected performed similarly (or better) than DA-7.
miRNA-regulated genes overlap analysis
Enrichment for Rb genes (Fig. 3 D) or differentiation genes (Fig. 4 B) among miRNA-regulated genes was performed according to Melton et al. (2010). In brief, the enrichment was calculated by the ratio between the observed number of overlapping transcripts and the expected number of overlapping transcripts, as: [genes in overlap of miRNA-regulated group and Rb genes/all genes in miRNA-regulated group]/[all Rb genes/all genes used in analysis to generate miRNA-regulated groups]. As previously observed by Melton et al. (2010), regulated gene sets (either miRNA-regulated or target gene sets) are frequently enriched for highly expressed transcripts. Thus, to avoid a bias in the analysis, only highly expressed transcripts were used in the overlap (defined as those above the 50th percentile of average raw signal of all the Affymetrix expression profiles). A similar pattern of results was obtained using all expressed transcripts, although all comparisons appear more highly enriched.
miRNA–mRNA interaction analysis on E1A regulated genes
We searched for DA-7 miRNA target genes (reported in Table S6) in the list of E1A-regulated genes. To account for cotargeting of DA-7 miRNAs and to achieve enough statistical power, the analysis was performed using the union of all DA-7 miRNA targets. These targets were stratified according to the cumulative number of MREs for DA-7 miRNAs: 1 or more (313 genes), 2 or more (173), 3 or more (101), and 4 or more (50). Then, we compared the regulation by E1A of genes in the background list (“No target”) with that of genes targeted by DA-7 miRNAs (Fig. 9 B). We considered the following gene classes: E1A up-regulated genes, further stratified in Rb-dependent and Rb-independent genes. Statistical significance of the target distribution was calculated in JMP9 (SAS) with one-way analysis of variance (ANOVA; P < 0.001) and the Dunnett’s t test (P < 0.05) as a post test, using the “No target” gene list as control group. Because the number of MREs usually correlates with the length of the 3′ UTR region, the correlation of gene expression data with the number of MREs for DA-7 was also adjusted for the length of 3′ UTR regions by Multivariate Linear Model within JMP9.
Clustering and statistical analyses
Heat maps were generated by Java TreeView software for Mac OS X. Statistical analyses were performed within the statistical software JMP9. Significance of the differences was calculated using ANOVA (in the case of more than two groups) or Welch’s t test. Categorical data were analyzed with a contingency analysis within JMP (SAS). P-values were calculated with a Fisher’s exact test.
Online supplemental material
Fig. S1 and S2 show the E1A-TD mRNA signature (GEX-MSC) and miRNA signature (MSC-miR) in MSCs, respectively. Fig. S3 shows that miRNA expression regulation during differentiation and upon E1A expression occurs at a transcriptional level. Fig. S4 shows the dose response effects of individual miRNA expression on proliferation and the combinatorial effects of DA-7 miRNAs on protein levels of differentiation and proliferation markers. Fig. S5 shows the enrichment analysis of DA-7 miRNA targets within the G1/S checkpoint pathway and the expression (as average copies per cell) of DA-7 miRNA families upon differentiation and E1A expression. Table S1 contains the list of primers used in the luciferase reporter assay. Table S2 contains details of the functional analysis performed on gene expression datasets. Table S3 contains details of the TRANSFAC analysis. Table S4 contains details of the E1A-TD miRNA signature. Table S5 contains details of miRNA– mRNA interactions within the Rb genes. Table S6 shows the list of miR targets. Table S7 contains details of the validation of miRNA–mRNA interactions for the G1/S checkpoint pathway. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201206033/DC1.
Acknowledgments
We are indebted to Bruno Amati and Heiko Muller for providing Myc ChIP-seq data; A. Berns and M. Vooijs for the Rb-loxP mice; Davide Cittaro, Giovanni d’Ario, and Barbara Felice for help with ChIP-seq data management and statistical and gene expression analyses; Pascale R. Romano for critically editing the manuscript; and Chiara Tordonato and Francesca Montani for help with immunofluorescence experiments.
This work was supported by grants from the Associazione Italiana per la Ricerca sul Cancro (AIRC), the Italian Ministries of Education University Research (MIUR), the European Community, the European Research Council, the Ferrari Foundation, and the Monzino Foundation to P.P. Di Fiore.
Footnotes
Abbreviations used in this paper:
- ANOVA
- analysis of variance
- CDS
- coding sequence
- ChIP
- chromatin immunoprecipitation
- ChIP-seq
- ChIP sequencing
- DA-miRNA
- differentiation-associated miRNA
- FDR
- false discovery rate
- GO
- gene ontology
- GSEA
- gene set enrichment analysis
- miRNA
- microRNA
- MRE
- miRNA responsive element
- MOI
- multiplicity of infection
- MSC
- muscle satellite cell
- MyHC
- myosin heavy chain
- NES
- normalized enrichment score
- PA-miRNA
- proliferation-associated miRNA
- Rb
- Retinoblastoma protein
- TD
- terminally differentiated
- TF
- transcription factor
- TSS
- transcription start sites
- UTR
- untranslated region
References
- Amati B., Alevizopoulos K., Vlach J. 1998. Myc and the cell cycle. Front. Biosci. 3:d250–d268 [DOI] [PubMed] [Google Scholar]
- Baek D., Villén J., Shin C., Camargo F.D., Gygi S.P., Bartel D.P. 2008. The impact of microRNAs on protein output. Nature. 455:64–71 10.1038/nature07242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. 2009. MicroRNAs: target recognition and regulatory functions. Cell. 136:215–233 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi F., Nuciforo P., Vecchi M., Bernard L., Tizzoni L., Marchetti A., Buttitta F., Felicioni L., Nicassio F., Di Fiore P.P. 2007. Survival prediction of stage I lung adenocarcinomas by expression of 10 genes. J. Clin. Invest. 117:3436–3444 10.1172/JCI32007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissels U., Wild S., Tomiuk S., Hafner M., Scheel H., Mihailovic A., Choi Y.H., Tuschl T., Bosio A. 2011. Combined characterization of microRNA and mRNA profiles delineates early differentiation pathways of CD133+ and CD34+ hematopoietic stem and progenitor cells. Stem Cells. 29:847–857 10.1002/stem.627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueno M.J., Gómez de Cedrón M., Laresgoiti U., Fernández-Piqueras J., Zubiaga A.M., Malumbres M. 2010. Multiple E2F-induced microRNAs prevent replicative stress in response to mitogenic signaling. Mol. Cell. Biol. 30:2983–2995 10.1128/MCB.01372-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushati N., Cohen S.M. 2007. microRNA functions. Annu. Rev. Cell Dev. Biol. 23:175–205 10.1146/annurev.cellbio.23.090506.123406 [DOI] [PubMed] [Google Scholar]
- Calin G.A., Croce C.M. 2006. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 6:857–866 10.1038/nrc1997 [DOI] [PubMed] [Google Scholar]
- Camarda G., Siepi F., Pajalunga D., Bernardini C., Rossi R., Montecucco A., Meccia E., Crescenzi M. 2004. A pRb-independent mechanism preserves the postmitotic state in terminally differentiated skeletal muscle cells. J. Cell Biol. 167:417–423 10.1083/jcb.200408164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannell I.G., Kong Y.W., Johnston S.J., Chen M.L., Collins H.M., Dobbyn H.C., Elia A., Kress T.R., Dickens M., Clemens M.J., et al. 2010. p38 MAPK/MK2-mediated induction of miR-34c following DNA damage prevents Myc-dependent DNA replication. Proc. Natl. Acad. Sci. USA. 107:5375–5380 10.1073/pnas.0910015107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Kumar R.M., Penn B.H., Berkes C.A., Kooperberg C., Boyer L.A., Young R.A., Tapscott S.J. 2006. Global and gene-specific analyses show distinct roles for Myod and Myog at a common set of promoters. EMBO J. 25:502–511 10.1038/sj.emboj.7600958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Yao Z., Sarkar D., Lawrence M., Sanchez G.J., Parker M.H., MacQuarrie K.L., Davison J., Morgan M.T., Ruzzo W.L., et al. 2010. Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Dev. Cell. 18:662–674 10.1016/j.devcel.2010.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T.C., Yu D., Lee Y.S., Wentzel E.A., Arking D.E., West K.M., Dang C.V., Thomas-Tikhonenko A., Mendell J.T. 2008. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat. Genet. 40:43–50 10.1038/ng.2007.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.F., Mandel E.M., Thomson J.M., Wu Q., Callis T.E., Hammond S.M., Conlon F.L., Wang D.Z. 2006. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 38:228–233 10.1038/ng1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicas A., Wang X., Zhang C., McCurrach M., Zhao Z., Mert O., Dickins R.A., Narita M., Zhang M., Lowe S.W. 2010. Dissecting the unique role of the retinoblastoma tumor suppressor during cellular senescence. Cancer Cell. 17:376–387 10.1016/j.ccr.2010.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crescenzi M., Soddu S., Sacchi A., Tatò F. 1995. Adenovirus infection induces reentry into the cell cycle of terminally differentiated skeletal muscle cells. Ann. N. Y. Acad. Sci. 752:9–18 10.1111/j.1749-6632.1995.tb17402.x [DOI] [PubMed] [Google Scholar]
- Croce C.M. 2008. Oncogenes and cancer. N. Engl. J. Med. 358:502–511 10.1056/NEJMra072367 [DOI] [PubMed] [Google Scholar]
- Easow G., Teleman A.A., Cohen S.M. 2007. Isolation of microRNA targets by miRNP immunopurification. RNA. 13:1198–1204 10.1261/rna.563707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari R., Pellegrini M., Horwitz G.A., Xie W., Berk A.J., Kurdistani S.K. 2008. Epigenetic reprogramming by adenovirus e1a. Science. 321:1086–1088 10.1126/science.1155546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P., Tchernyshyov I., Chang T.C., Lee Y.S., Kita K., Ochi T., Zeller K.I., De Marzo A.M., Van Eyk J.E., Mendell J.T., Dang C.V. 2009. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 458:762–765 10.1038/nature07823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandori C., Cowley S.M., James L.P., Eisenman R.N. 2000. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell Dev. Biol. 16:653–699 10.1146/annurev.cellbio.16.1.653 [DOI] [PubMed] [Google Scholar]
- Guo H., Ingolia N.T., Weissman J.S., Bartel D.P. 2010. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 466:835–840 10.1038/nature09267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R.A. 2011. Hallmarks of cancer: the next generation. Cell. 144:646–674 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- He L., He X., Lim L.P., de Stanchina E., Xuan Z., Liang Y., Xue W., Zender L., Magnus J., Ridzon D., et al. 2007. A microRNA component of the p53 tumour suppressor network. Nature. 447:1130–1134 10.1038/nature05939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Kasinski A.L., Slack F.J. 2011. MicroRNA therapeutics in preclinical cancer models. Lancet Oncol. 12:319–321 10.1016/S1470-2045(11)70067-5 [DOI] [PubMed] [Google Scholar]
- Kloosterman W.P., Plasterk R.H. 2006. The diverse functions of microRNAs in animal development and disease. Dev. Cell. 11:441–450 10.1016/j.devcel.2006.09.009 [DOI] [PubMed] [Google Scholar]
- Kota J., Chivukula R.R., O’Donnell K.A., Wentzel E.A., Montgomery C.L., Hwang H.W., Chang T.C., Vivekanandan P., Torbenson M., Clark K.R., et al. 2009. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 137:1005–1017 10.1016/j.cell.2009.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J., Loedige I., Filipowicz W. 2010. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 11:597–610 [DOI] [PubMed] [Google Scholar]
- Kumar M.S., Lu J., Mercer K.L., Golub T.R., Jacks T. 2007. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat. Genet. 39:673–677 10.1038/ng2003 [DOI] [PubMed] [Google Scholar]
- Kumar M.S., Erkeland S.J., Pester R.E., Chen C.Y., Ebert M.S., Sharp P.A., Jacks T. 2008. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc. Natl. Acad. Sci. USA. 105:3903–3908 10.1073/pnas.0712321105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M.S., Pester R.E., Chen C.Y., Lane K., Chin C., Lu J., Kirsch D.G., Golub T.R., Jacks T. 2009. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 23:2700–2704 10.1101/gad.1848209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf P., Rusu M., Sheridan R., Sewer A., Iovino N., Aravin A., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M., et al. 2007. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 129:1401–1414 10.1016/j.cell.2007.04.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B.P., Burge C.B., Bartel D.P. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 120:15–20 10.1016/j.cell.2004.12.035 [DOI] [PubMed] [Google Scholar]
- Lipinski M.M., Jacks T. 1999. The retinoblastoma gene family in differentiation and development. Oncogene. 18:7873–7882 10.1038/sj.onc.1203244 [DOI] [PubMed] [Google Scholar]
- Liu C., Kelnar K., Liu B., Chen X., Calhoun-Davis T., Li H., Patrawala L., Yan H., Jeter C., Honorio S., et al. 2011. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat. Med. 17:211–215 10.1038/nm.2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Getz G., Miska E.A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A., et al. 2005. MicroRNA expression profiles classify human cancers. Nature. 435:834–838 10.1038/nature03702 [DOI] [PubMed] [Google Scholar]
- Marasa B.S., Srikantan S., Masuda K., Abdelmohsen K., Kuwano Y., Yang X., Martindale J.L., Rinker-Schaeffer C.W., Gorospe M. 2009. Increased MKK4 abundance with replicative senescence is linked to the joint reduction of multiple microRNAs. Sci. Signal. 2:ra69 10.1126/scisignal.2000442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A., Levine S.S., Cole M.F., Frampton G.M., Brambrink T., Johnstone S., Guenther M.G., Johnston W.K., Wernig M., Newman J., et al. 2008. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 134:521–533 10.1016/j.cell.2008.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martello G., Rosato A., Ferrari F., Manfrin A., Cordenonsi M., Dupont S., Enzo E., Guzzardo V., Rondina M., Spruce T., et al. 2010. A MicroRNA targeting dicer for metastasis control. Cell. 141:1195–1207 10.1016/j.cell.2010.05.017 [DOI] [PubMed] [Google Scholar]
- Melton C., Judson R.L., Blelloch R. 2010. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 463:621–626 10.1038/nature08725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt W.M., Lin Y.G., Han L.Y., Kamat A.A., Spannuth W.A., Schmandt R., Urbauer D., Pennacchio L.A., Cheng J.F., Nick A.M., et al. 2008. Dicer, Drosha, and outcomes in patients with ovarian cancer. N. Engl. J. Med. 359:2641–2650 10.1056/NEJMoa0803785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestdagh P., Lefever S., Pattyn F., Ridzon D., Fredlund E., Fieuw A., Ongenaert M., Vermeulen J., De Paepe A., Wong L., et al. 2011. The microRNA body map: dissecting microRNA function through integrative genomics. Nucleic Acids Res. 39:e136 10.1093/nar/gkr646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicassio F., Bianchi F., Capra M., Vecchi M., Confalonieri S., Bianchi M., Pajalunga D., Crescenzi M., Bonapace I.M., Di Fiore P.P. 2005. A cancer-specific transcriptional signature in human neoplasia. J. Clin. Invest. 115:3015–3025 10.1172/JCI24862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell K.A., Wentzel E.A., Zeller K.I., Dang C.V., Mendell J.T. 2005. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 435:839–843 10.1038/nature03677 [DOI] [PubMed] [Google Scholar]
- Ozen M., Creighton C.J., Ozdemir M., Ittmann M. 2008. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene. 27:1788–1793 10.1038/sj.onc.1210809 [DOI] [PubMed] [Google Scholar]
- Perna D., Fagà G., Verrecchia A., Gorski M.M., Barozzi I., Narang V., Khng J., Lim K.C., Sung W.K., Sanges R., et al. 2012. Genome-wide mapping of Myc binding and gene regulation in serum-stimulated fibroblasts. Oncogene. 31:1695–1709 10.1038/onc.2011.359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrello A., Cerone M.A., Coen S., Gurtner A., Fontemaggi G., Cimino L., Piaggio G., Sacchi A., Soddu S. 2000. p53 regulates myogenesis by triggering the differentiation activity of pRb. J. Cell Biol. 151:1295–1304 10.1083/jcb.151.6.1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri P.L., Avantaggiati M.L., Balsano C., Sang N., Graessmann A., Giordano A., Levrero M. 1997. p300 is required for MyoD-dependent cell cycle arrest and muscle-specific gene transcription. EMBO J. 16:369–383 10.1093/emboj/16.2.369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao P.K., Kumar R.M., Farkhondeh M., Baskerville S., Lodish H.F. 2006. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc. Natl. Acad. Sci. USA. 103:8721–8726 10.1073/pnas.0602831103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart B.J., Slack F.J., Basson M., Pasquinelli A.E., Bettinger J.C., Rougvie A.E., Horvitz H.R., Ruvkun G. 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 403:901–906 10.1038/35002607 [DOI] [PubMed] [Google Scholar]
- Sachdeva M., Zhu S., Wu F., Wu H., Walia V., Kumar S., Elble R., Watabe K., Mo Y.Y. 2009. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc. Natl. Acad. Sci. USA. 106:3207–3212 10.1073/pnas.0808042106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M., Schwanhäusser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. 2008. Widespread changes in protein synthesis induced by microRNAs. Nature. 455:58–63 10.1038/nature07228 [DOI] [PubMed] [Google Scholar]
- Sethupathy P., Corda B., Hatzigeorgiou A.G. 2006. TarBase: A comprehensive database of experimentally supported animal microRNA targets. RNA. 12:192–197 10.1261/rna.2239606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr C.J. 1996. Cancer cell cycles. Science. 274:1672–1677 10.1126/science.274.5293.1672 [DOI] [PubMed] [Google Scholar]
- Small E.M., O’Rourke J.R., Moresi V., Sutherland L.B., McAnally J., Gerard R.D., Richardson J.A., Olson E.N. 2010. Regulation of PI3-kinase/Akt signaling by muscle-enriched microRNA-486. Proc. Natl. Acad. Sci. USA. 107:4218–4223 10.1073/pnas.1000300107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A., Brennecke J., Bushati N., Russell R.B., Cohen S.M. 2005. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3’UTR evolution. Cell. 123:1133–1146 10.1016/j.cell.2005.11.023 [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taulli R., Bersani F., Foglizzo V., Linari A., Vigna E., Ladanyi M., Tuschl T., Ponzetto C. 2009. The muscle-specific microRNA miR-206 blocks human rhabdomyosarcoma growth in xenotransplanted mice by promoting myogenic differentiation. J. Clin. Invest. 119:2366–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson J.M., Parker J., Perou C.M., Hammond S.M. 2004. A custom microarray platform for analysis of microRNA gene expression. Nat. Methods. 1:47–53 10.1038/nmeth704 [DOI] [PubMed] [Google Scholar]
- Tiainen M., Spitkovsky D., Jansen-Dürr P., Sacchi A., Crescenzi M. 1996. Expression of E1A in terminally differentiated muscle cells reactivates the cell cycle and suppresses tissue-specific genes by separable mechanisms. Mol. Cell. Biol. 16:5302–5312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting Y., Medina D.J., Strair R.K., Schaar D.G. 2010. Differentiation-associated miR-22 represses Max expression and inhibits cell cycle progression. Biochem. Biophys. Res. Commun. 394:606–611 10.1016/j.bbrc.2010.03.030 [DOI] [PubMed] [Google Scholar]
- Trang P., Wiggins J.F., Daige C.L., Cho C., Omotola M., Brown D., Weidhaas J.B., Bader A.G., Slack F.J. 2011. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol. Ther. 19:1116–1122 10.1038/mt.2011.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimarchi J.M., Lees J.A. 2002. Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell Biol. 3:11–20 10.1038/nrm714 [DOI] [PubMed] [Google Scholar]
- Tsang J.S., Ebert M.S., van Oudenaarden A. 2010. Genome-wide dissection of microRNA functions and cotargeting networks using gene set signatures. Mol. Cell. 38:140–153 10.1016/j.molcel.2010.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen S., Abreu-Goodger C., Enright A.J. 2008. Detecting microRNA binding and siRNA off-target effects from expression data. Nat. Methods. 5:1023–1025 10.1038/nmeth.1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Garzon R., Sun H., Ladner K.J., Singh R., Dahlman J., Cheng A., Hall B.M., Qualman S.J., Chandler D.S., et al. 2008. NF-kappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell. 14:369–381 10.1016/j.ccr.2008.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman B., Ha I., Ruvkun G. 1993. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 75:855–862 10.1016/0092-8674(93)90530-4 [DOI] [PubMed] [Google Scholar]
- Williams A.H., Liu N., van Rooij E., Olson E.N. 2009. MicroRNA control of muscle development and disease. Curr. Opin. Cell Biol. 21:461–469 10.1016/j.ceb.2009.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods K., Thomson J.M., Hammond S.M. 2007. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J. Biol. Chem. 282:2130–2134 10.1074/jbc.C600252200 [DOI] [PubMed] [Google Scholar]
- Wu W. 2011. Modulation of microRNAs for potential cancer therapeutics. Methods Mol. Biol. 676:59–70 10.1007/978-1-60761-863-8_5 [DOI] [PubMed] [Google Scholar]
- Xiao F., Zuo Z., Cai G., Kang S., Gao X., Li T. 2009. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res. 37:D105–D110 10.1093/nar/gkn851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeilding N.M., Procopio W.N., Rehman M.T., Lee W.M. 1998. c-myc mRNA is down-regulated during myogenic differentiation by accelerated decay that depends on translation of regulatory coding elements. J. Biol. Chem. 273:15749–15757 10.1074/jbc.273.25.15749 [DOI] [PubMed] [Google Scholar]