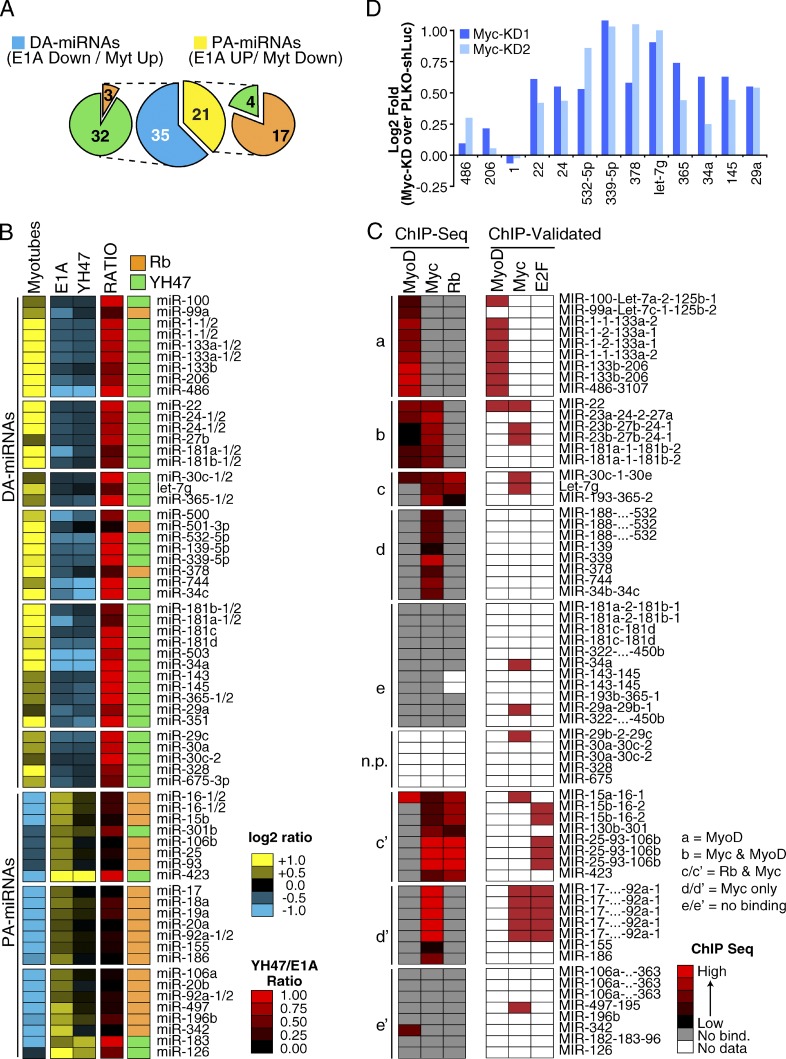
Figure 2.
E1A-TD miRNA signature (C2C12-miR). (A) A series of pie charts summarize the E1A-TD miRNA signature, which consists of differentiation-associated (DA-miRNAs, n = 35) and proliferation-associated (PA-miRNAs, n = 21) miRNAs. Each class is subsequently distinguished according to Rb pathway responsiveness (Rb, in orange; and YH47, in green). (B) The heat map shows the regulation of miRNAs in the E1A-TD signature. Mature miRNAs expressed from multiple genomic loci are also shown (i.e., miR-133a-1 and -133a-2) to allow direct comparison with ChIP-seq data. (C) Meta-analysis of the E1A-TD miRNA signature using ChIP-seq datasets. Rows show the normalized binding of each TF at the corresponding miRNA promoter. miRNA–TF interactions experimentally validated by direct ChIP experiments are also shown (“ChIP-Validated,” see Materials and methods for details). (D) miRNA regulation upon Myc knockdown in C2C12 myoblasts was evaluated through RT-qPCR. Two different shRNAs (PLKO-Myc-KD1 and KD2) were used. Results are normalized to myoblasts expressing an shRNA against luciferase (PLKO–shLuc). U5A was used as housekeeping gene. A group of MyoD-bound muscle-specific miRNAs (miR-1, -206, and -486) was used as a negative control. The data shown are from a single representative experiment out of two repeats.