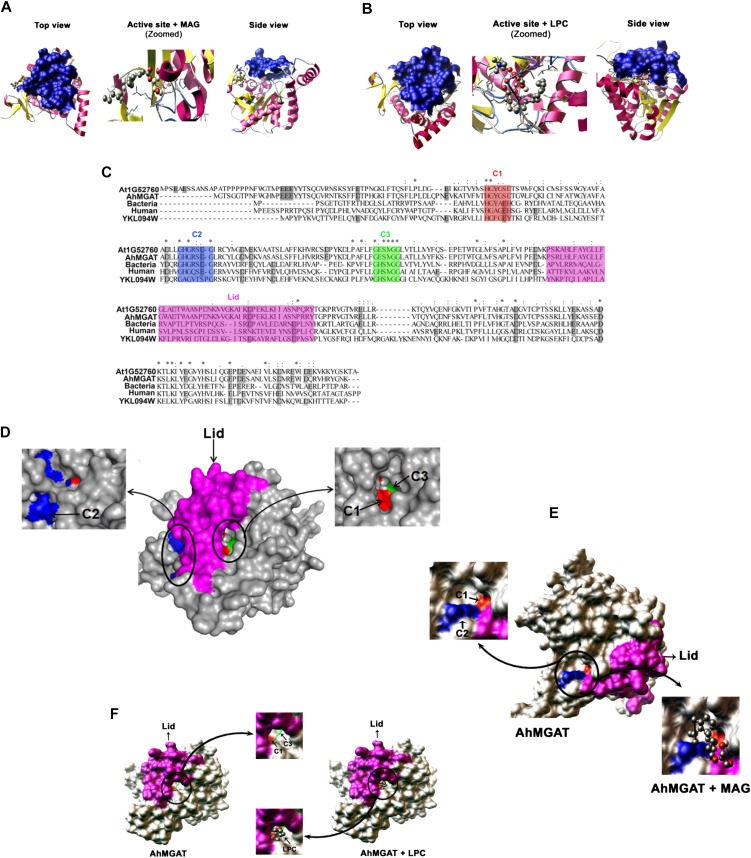
Figure 11.
Homology modeling and molecular docking studies on AhMGAT protein. A and B, The three-dimensional model for AhMGAT protein was built using a MAG lipase (Protein Data Bank code 3PE6) structure, and the quality of the homology model was assessed using a Ramachandran plot. The docking of lipid substrates demonstrated the binding orientations of MAG (1-oleoyl; A) and LPC (B) on AhMGAT protein. The snapshots of docking complexes (AhMGAT protein with MAG or LPC) are shown as top and side views. MAG and LPC substrates are shown in ball-and-stick representation. The lid structure that covers the LPC binding region is shown as a blue-colored surface. C, Multiple sequence alignment of AhMGAT protein and its homologs. The conserved regions are denoted as C1 (HXYXXD), C2 (GHGXSXG), and C3 (GXSXG) motifs. The color codes are as follows: C1, red; C2, blue; C3, green; lid, magenta. D, A surface view of AhMGAT illustrates the spatial organization of the conserved regions. C2 and C1 are located on the AhMGAT protein surface, and MAG forms a stable interaction with the binding free energy of −3.23 kcal mol−1. E, A surface view of AhMGAT protein structure revealed that C1 and C3 are closely arranged under the lid structure where the LPC binds (−4.81 kcal mol−1). F, The C1 region is located at an equal distance from the C2 and C3 regions.