Abstract
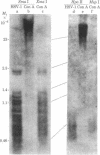
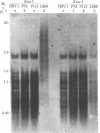
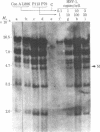
An in vitro model of latency of herpes simplex virus type 1 (HSV-1) in a lymphoid cell line has been developed recently. CEM cells persistently infected with HSV-1 transiently ceased to produce virus for 24 days. This nonproductive state could either be reversed with phytohemagglutinin or maintained with concanavalin A. This system was used to study the relationship between DNA methylation and HSV-1 latency. DNA was probed for methylation by comparing the cleavage patterns generated by two pairs of restriction endonucleases (Sma I vs. Xma I and Hpa II vs. MspI); these enzymes show differential activity reflecting methylation of the recognition sequences. Viral DNA in the concanavalin A-treated cells (not producing virus) was found to be extensively methylated. By contrast, no methylated copies were detected in viral DNA from producer cells. About 800 days after the initial infection, the productive culture once again became nonproductive. Viral sequences in the latter cells were also methylated. Reconstitution experiments revealed 1-2 copies of viral DNA in cells from the latent stages and 40-80 copies in cells from productive stages. Most (if not all) of the viral genome is present in cells from various productive and latent stages. No differences in sequence arrangement were detected (although a terminal fragment of intracellular HSV-1 DNA appeared to be under-represented in latent cells). These results suggest a role for DNA methylation in the mechanism of HSV-1 latency in this system.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bird A. P., Southern E. M. Use of restriction enzymes to study eukaryotic DNA methylation: I. The methylation pattern in ribosomal DNA from Xenopus laevis. J Mol Biol. 1978 Jan 5;118(1):27–47. doi: 10.1016/0022-2836(78)90242-5. [DOI] [PubMed] [Google Scholar]
- Cohen J. C. Methylation of milk-borne and genetically transmitted mouse mammary tumor virus proviral DNA. Cell. 1980 Mar;19(3):653–662. doi: 10.1016/s0092-8674(80)80042-0. [DOI] [PubMed] [Google Scholar]
- Compere S. J., Palmiter R. D. DNA methylation controls the inducibility of the mouse metallothionein-I gene lymphoid cells. Cell. 1981 Jul;25(1):233–240. doi: 10.1016/0092-8674(81)90248-8. [DOI] [PubMed] [Google Scholar]
- Desrosiers R. C., Mulder C., Fleckenstein B. Methylation of Herpesvirus saimiri DNA in lymphoid tumor cell lines. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3839–3843. doi: 10.1073/pnas.76.8.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M., Wang R. Y. 5-Methylcytosine in eukaryotic DNA. Science. 1981 Jun 19;212(4501):1350–1357. doi: 10.1126/science.6262918. [DOI] [PubMed] [Google Scholar]
- Goldin A. L., Sandri-Goldin R. M., Levine M., Glorioso J. C. Cloning of herpes simplex virus type 1 sequences representing the whole genome. J Virol. 1981 Apr;38(1):50–58. doi: 10.1128/jvi.38.1.50-58.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groudine M., Eisenman R., Weintraub H. Chromatin structure of endogenous retroviral genes and activation by an inhibitor of DNA methylation. Nature. 1981 Jul 23;292(5821):311–317. doi: 10.1038/292311a0. [DOI] [PubMed] [Google Scholar]
- Hammer S. M., Richter B. S., Hirsch M. S. Activation and suppression of herpes simplex virus in a human T lymphoid cell line. J Immunol. 1981 Jul;127(1):144–148. [PubMed] [Google Scholar]
- Lindahl T., Adams A., Bjursell G., Bornkamm G. W., Kaschka-Dierich C., Jehn U. Covalently closed circular duplex DNA of Epstein-Barr virus in a human lymphoid cell line. J Mol Biol. 1976 Apr 15;102(3):511–530. doi: 10.1016/0022-2836(76)90331-4. [DOI] [PubMed] [Google Scholar]
- Mandel J. L., Chambon P. DNA methylation: organ specific variations in the methylation pattern within and around ovalbumin and other chicken genes. Nucleic Acids Res. 1979 Dec 20;7(8):2081–2103. doi: 10.1093/nar/7.8.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee J. D., Ginder G. D. Specific DNA methylation sites in the vicinity of the chicken beta-globin genes. Nature. 1979 Aug 2;280(5721):419–420. doi: 10.1038/280419a0. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Skare J., Summers W. C. Structure and function of herpesvirus genomes. II. EcoRl, Sbal, and HindIII endonuclease cleavage sites on herpes simplex virus. Virology. 1977 Feb;76(2):581–595. doi: 10.1016/0042-6822(77)90240-9. [DOI] [PubMed] [Google Scholar]
- Sutter D., Doerfler W. Methylation of integrated adenovirus type 12 DNA sequences in transformed cells is inversely correlated with viral gene expression. Proc Natl Acad Sci U S A. 1980 Jan;77(1):253–256. doi: 10.1073/pnas.77.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waalwijk C., Flavell R. A. DNA methylation at a CCGG sequence in the large intron of the rabbit beta-globin gene: tissue-specific variations. Nucleic Acids Res. 1978 Dec;5(12):4631–4634. doi: 10.1093/nar/5.12.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waalwijk C., Flavell R. A. MspI, an isoschizomer of hpaII which cleaves both unmethylated and methylated hpaII sites. Nucleic Acids Res. 1978 Sep;5(9):3231–3236. doi: 10.1093/nar/5.9.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner F. J., Bornkamm G. W., Fleckenstein B. Episomal viral DNA in a Herpesvirus saimiri-transformed lymphoid cell line. J Virol. 1977 Jun;22(3):794–803. doi: 10.1128/jvi.22.3.794-803.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi M., Koshland M. E. Expression of the J chain gene during B cell differentiation is inversely correlated with DNA methylation. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4907–4911. doi: 10.1073/pnas.78.8.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssoufian H., Mulder C. Detection of methylated sequences in eukaryotic DNA with the restriction endonucleases Smai and Xmai. J Mol Biol. 1981 Jul 25;150(1):133–136. doi: 10.1016/0022-2836(81)90328-4. [DOI] [PubMed] [Google Scholar]
- van der Ploeg L. H., Flavell R. A. DNA methylation in the human gamma delta beta-globin locus in erythroid and nonerythroid tissues. Cell. 1980 Apr;19(4):947–958. doi: 10.1016/0092-8674(80)90086-0. [DOI] [PubMed] [Google Scholar]