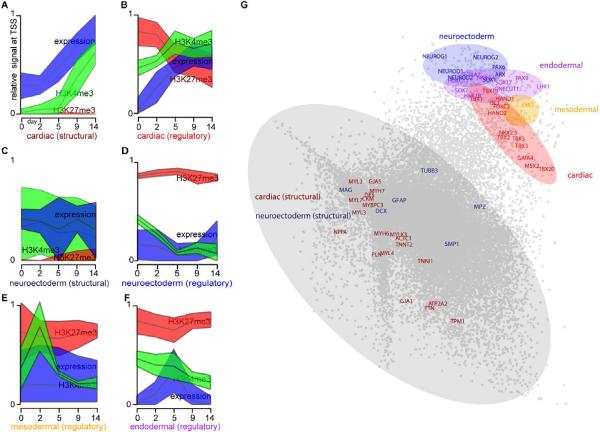
Figure 3. Temporal chromatin signatures enable cross-lineage identification of key regulatory factors.
(A–F) The median levels (middle line) and 95% confidence intervals (shaded regions) of H3K27me3 (red), H3K4me3 (green) and RNA expression (blue) at each time point are depicted for several categories of genes: (A) genes involved in cardiac structure and function, (C) genes involved in neuroectoderm structure/function, and regulators of differentiation for (B) cardiac, (D) neuroectoderm, (E) mesoodermal and (F) endodermal cells. (A–F) were identically normalized, such that the lowest and highest values for each individual mark across all time points and gene groups were plotted as 0 and 1, respectively. (G) Using principle component analysis, the 15 dimensional data for each gene (5 time points * 3 measurements of chromatin and mRNA) were reduced to two dimensions, and a scatterplot is shown depicted the relative locations of each gene in the reduced-dimensional space. Genes involved in structure and function of cells are contained within the largest cluster (grey) distinct from the cluster containing key regulators of cellular fate: cardiac (red), neuroectoderm (blue), endoderm (purple) and mesoderm (yellow). Non-annotated genes within each of the colored domains have a high probability of having unappreciated roles as key regulators of cellular fate.