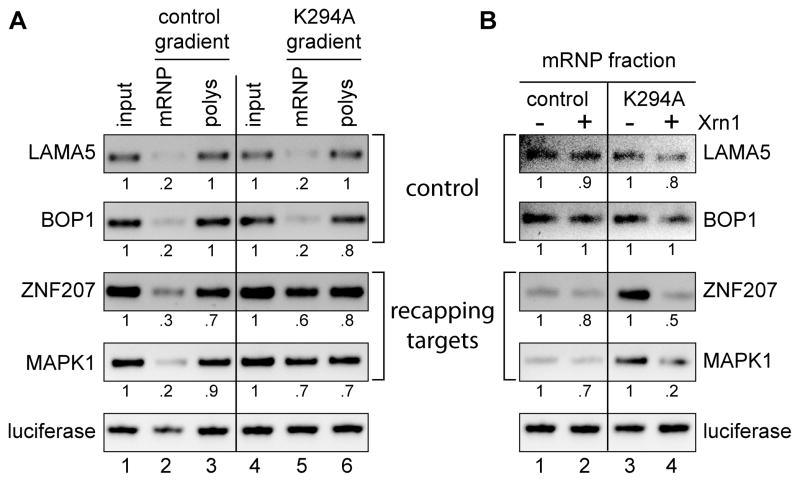
Figure 6. The recapping targets that accumulate in non-translating mRNP are uncapped.
(A) RNA was recovered from each of the input cytoplasmic extracts from Figure 5 (input), and pooled mRNP (1,3,5) and polysome fractions (13,15,17,19,21) from each gradient. Each pool received an equal amount of dephosphorylated firefly luciferase RNA as an internal control prior to RNA purification, which was followed by cDNA synthesis, and the cDNAs were analyzed by semi-quantitative RT-PCR using the same 3′-weighted primers as in Figure 5(B and C). The products were separated on a 1.2% agarose gel and quantified using a BioRad GelDoc Imager and Quantity One 1-D gel analysis software. Results were normalized to the internal luciferase control. (B) Sucrose step gradients were used to recover mRNP fractions from triplicate cultures of control and K294A-expressing cells. An equal amount of dephosphorylated firefly luciferase RNA was added to each sample as an Xrn1-resistant control and the recovered RNA was treated Xrn1 to degrade uncapped transcripts. Individual samples were amplified by semi-quantitative RT-PCR using primers located near the 5′ ends of two recapping targets (ZNF207, MAPK1) and two controls (BOP1, LAMA5), the pooled products were separated on a 1.2% agarose gel and visualized and quantified as in (A).