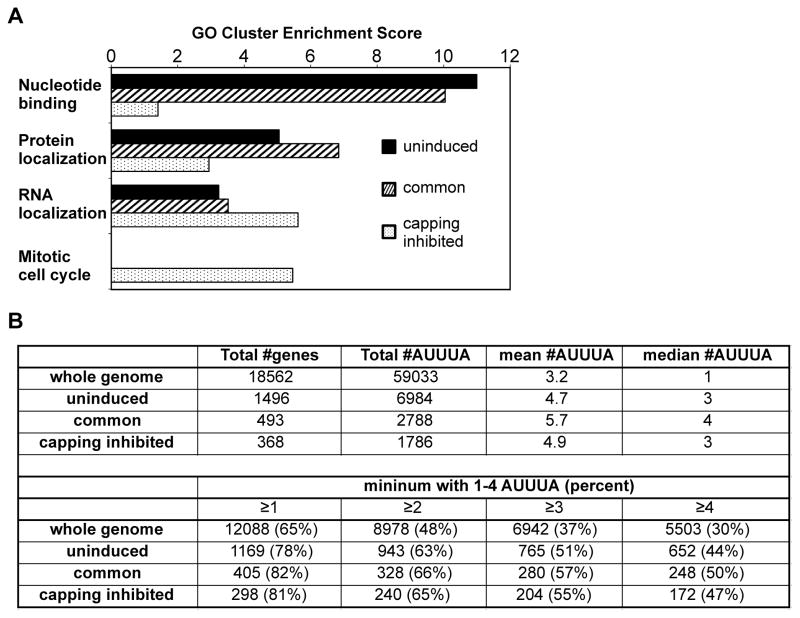
Figure 7. Properties of recapping targets.
(A) Gene ontology analysis of transcripts in the uninduced, common and capping inhibited transcripts sets was performed using the NIH DAVID tool with default settings and high stringency. The top 4 cluster groupings are shown as a function of each of the transcript sets, with additional groupings in Supplemental Table S3. (B) A python script was developed to search the AREsite database for AUUUA elements present in the 3′-UTRs of the genes that correspond to the 55,662 Ensembl transcripts that were analyzed in Figure 1. The upper panel details the total number of AUUUA elements and the mean and median number of elements per transcript in each dataset, and in the lower panel these are broken down by the percentage of transcripts with different numbers of AUUUA elements.