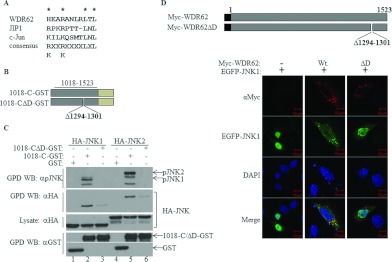
Figure 4. A putative D domain within WDR62 is responsible for WDR62–JNK association.
(A) Alignment of the amino acid sequences of the D domains derived from WDR62, JIP1 and c-Jun. The consensus D domain sequence identified in numerous proteins is shown. (B) Schematic representation of the WDR62 deletion constructs used in the experiments. Amino acid positions are numbered. The light green rectangle represents the position of the GST tag. (C) HEK-293T cells were transfected with plasmids encoding GST, 1018-C–GST or 1018-CΔD–GST together with HA–JNK1/2 as indicated. GST-containing complexes were isolated from cell lysates with glutathione–agarose beads, washed extensively and eluted with reduced glutathione. The protein complexes were subjected to Western blotting (WB) with anti-pJNK (top panel), anti-HA (second panel from the top) or anti-GST (bottom panel) antibodies. The expression level of HA–JNK1/2 was determined by blotting the total cell lysate with anti-HA antibody (middle panel). The migration of the relevant proteins is indicated by arrows. (D) Top panel: schematic representation of the full-length WDR62 wild-type and deletion mutant constructs used in the HeLa cell immunofluorescence staining experiments. Amino acid positions are numbered. The black square represents the position of the Myc epitope tag. Bottom panels: HeLa cells were co-transfected with an EGFP–JNK1 expression plasmid together with plasmids encoding either wild-type WDR62 (Wt) or the D domain deletion mutant (ΔD). Cells were fixed and stained with anti-Myc antibody (red). EGFP–JNK1 is shown in green. Co-localization appears in yellow in the merged image. Nuclei were stained with DAPI (blue). Representative confocal microscopy images are shown. Scale bar, 10 μm. GPD, GST pull-down.