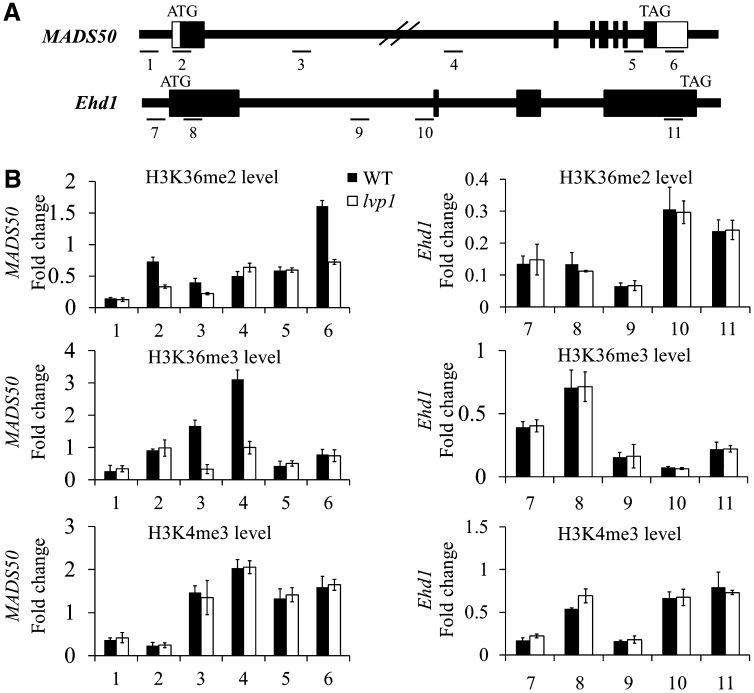
Figure 6.
H3K36me2/3 Levels Are Reduced at the MADS50 Locus in lvp1 Mutant Plants.
ChIP samples were analyzed by quantitative PCR of six regions of MADS50 and five regions of Ehd1. The rice Actin1 gene was used as an internal control. The relative levels of specific histone H3 Lys methylation marks were calculated as the ratio of specific methylation marks over total H3 and normalized as the ratio over total H3 at Actin1; error bars indicate sd (n = 3 or more). Primers are listed in Supplemental Table 1 online.
(A) Genomic structure of the MADS50 and Ehd1 loci. Regions assayed in (B) are indicated by lines and numbered.
(B) ChIP analysis of H3K36me2/3 and H3K4me3 deposited on the chromatin of MADS50 and Ehd1 loci in wild-type (WT) and lvp1 mutant plants.