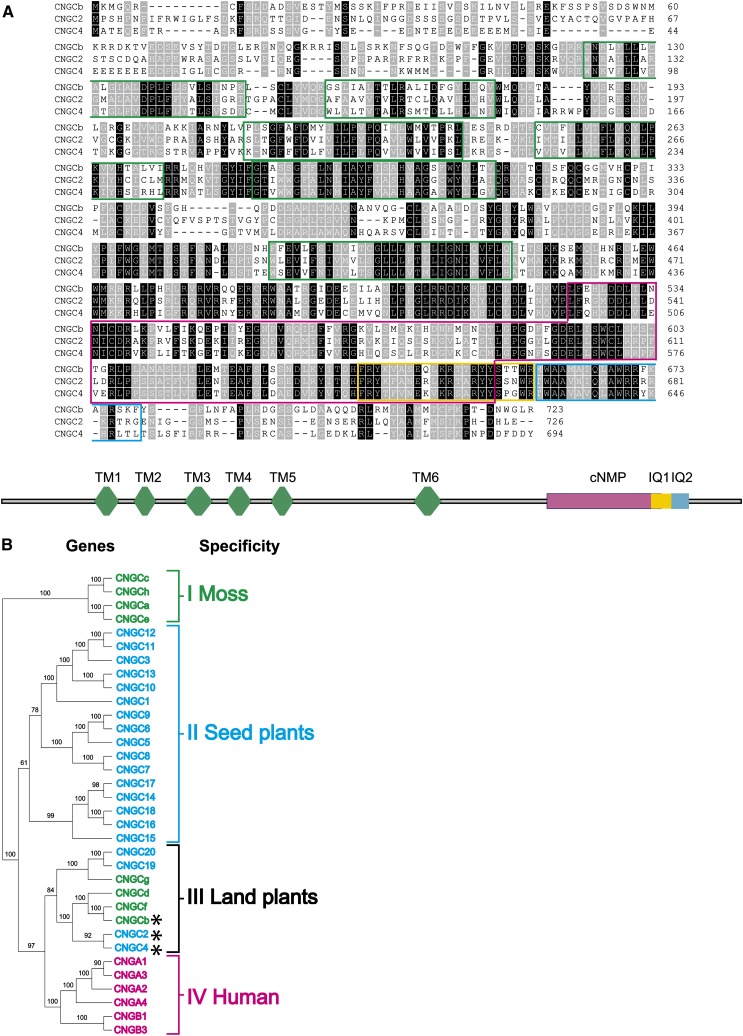
Figure 1.
Phylogenetic and Structural Organization of Moss and Arabidopsis CNGCs.
(A) Alignment of the deduced amino acid sequences of moss CNGCb with Arabidopsis CNGC2 and CNGC4. Black background denotes identical residues and gray background similar residues. The six predicted transmembrane helices are delimited by green boxes, the cyclic nucleotide binding domain by a red box, the canonical calmodulin binding domains (IQ1) by a yellow box, and the second noncanonical calmodulin binding domain (IQ2) by a blue box.
(B) Plant and human amino acid sequences of CNGCs (see Supplemental Data Set 1 online) are compared in an unrooted neighbor-joining tree (see Methods). The tree shows four main gene clusters that are significantly different: cluster I (green), containing only moss-specific CNGCs; cluster II (blue), containing only plant specific CNGCs; cluster III (black), containing both moss and seed plant CNGCs and is therefore inferred to be land plant specific; and cluster IV (magenta), containing only human CNGCs. The CNGCb, CNGC2, and CNGC4 that presumably encode thermosensory Ca2+channels in land plants are marked with asterisks.