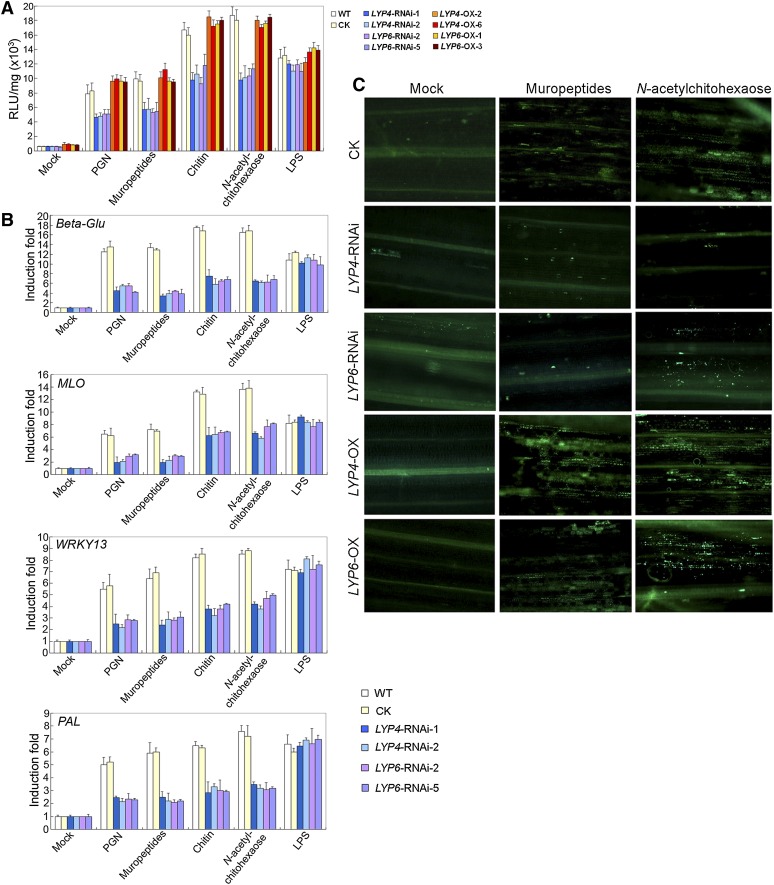
Figure 3.
RNAi Silencing or Overexpressing of LYP4 and LYP6 Specifically Modulate PGN- and Chitin- but Not LPS-Induced Defense Responses in Rice.
(A) ROS generation induced by PGN or chitin is significantly suppressed in LYP4 or LYP6 RNAi transgenic rice lines. Roots from 4-d-old wild-type or transgenic rice seedlings were incubated with 100 μg/mL PGNXoo, PGNXoo muropeptides, crab shell chitin, N-acetylchitohexaose, LPS, or sterile water (mock) for 30 min before ROS measurement. The data represent the mean ± sd of nine samples from three independent tests. CK, control (empty vector transgenic rice); RLU, relative light units; WT, the wild type.
(B) Defense gene activation induced by PGN or chitin is compromised in LYP4 or LYP6 RNAi transgenic rice lines. Callus cells from wild-type or RNAi transgenic rice lines were incubated with 100 μg/mL PGNXoo, PGNXoo muropeptides, crab shell chitin, N-acetylchitohexaose, LPS, or sterile water (mock) for 30 min, and the induction of four representative defense marker genes Beta-Glu, MLO, WRKY13, and PAL was determined by qPCR. The data represent the mean ± sd of nine samples from three independent tests.
(C) Callose deposition induced by PGN or chitin is substantially impaired in LYP4 and LYP6 RNAi transgenic rice but enhanced in LYP OX rice. The first leaf of 5-d-old wild-type, LYP4-RNAi-1, LYP6-RNAi-2, LYP4-OX-2, or LYP6-OX-3 rice seedling was immersed into solution containing 10 μg/mL PGNXoo muropeptides or N-acetylchitohexaose and vacuumed for 30 min. Callose staining was conducted 18 h later.
At least three biological repeats were conducted for individual experiments.