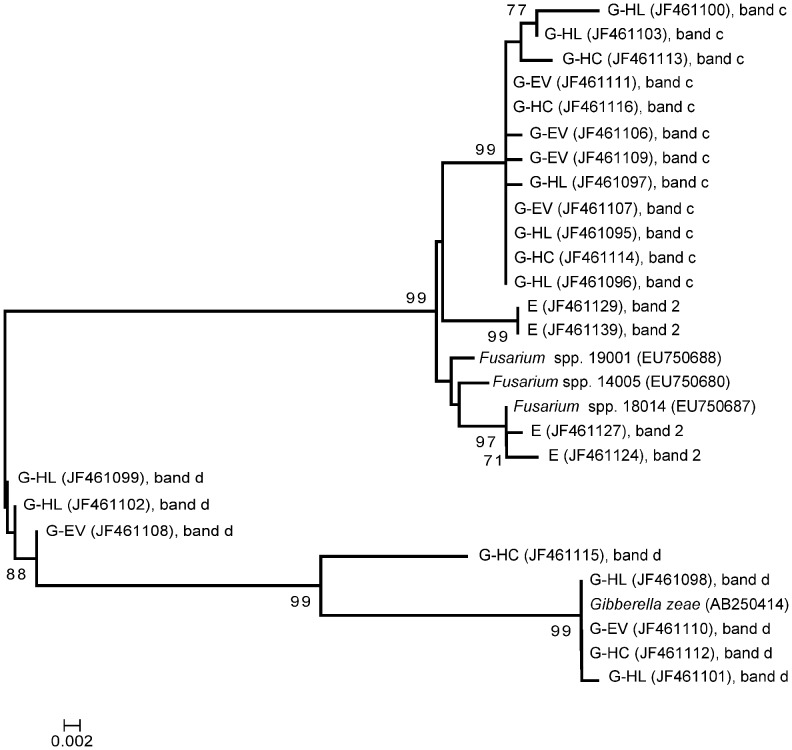
Figure 3. Neighbor-joining tree derived from ITS sequences amplified from TC DNA extracted from surface sterilized eggs (E) and gut of WCR larvae feeding on maize plants grown in Haplic Chernozem (G-HC), in Haplic Luvisol (G-HL) and in Eutric Vertisol (G-EV).
Each sequence is labeled with the corresponding GenBank accession number, and the corresponding DGGE band in Fig. 1 and 2. The dendrogram was generated with MEGA 4 software. The branches show bootstrap values higher than 60%.