Abstract
Targeting the epidermal growth factor receptor (EGFR) using the tyrosine kinase inhibitor (TKI) erlotinib has demonstrated activity in aerodigestive tract malignancies. Co-targeting of the G-protein-coupled receptor cyclooxygenase (COX) with EGFR inhibitors has shown promise in preclinical models and early phase clinical studies. We studied the modulation of serum proteins after neoadjuvant treatment with erlotinib with or without sulindac in head and neck cancer patients. In a prospective, randomized, double-blind clinical trial, paired serum samples were obtained before and after neoadjuvant treatment in three groups of patients (n=23 total), who were randomized to receive 7 - 14 consecutive days of erlotinib alone, erlotinib plus sulindac, or placebo. Two separate multiplexed ELISA systems (SearchLight™ or Luminex™) were used to measure serum biomarkers. HGF and IL-6 levels were tested on both systems, and validated using single analyte ELISAs. Several analytes were significantly altered (generally decreased) post-treatment, in patients who received erlotinib (with or without sulindac) as well as in the placebo groups. No single analyte was differentially altered across the three treatment groups using either multiplex platform. Single HGF ELISA suggested a nonspecific decrease in all patients. These results demonstrate the importance of a placebo group when assessing changes in expression of serum biomarkers. While multiplex platforms can provide quantitative information on a large number of serum analytes, results should be cautiously compared across platforms due to their intrinsic features. Furthermore, the dynamic range of expression of a single analyte is constrained in multiplex versus standard ELISA.
Keywords: HNSCC, Biomarkers, EGFR, COX, Erlotinib, Sulindac
Introduction
Increased understanding of the molecular alterations in head and neck squamous cell carcinoma (HNSCC) has led to augmented efforts to develop molecular targeted therapies (1). An increasingly common target is the epidermal growth factor receptor (EGFR) a transmembrane protein vital for the proliferation, survival, invasion and angiogenesis. Small-molecule EGFR-specific tyrosine kinase inhibitors (TKI), such as erlotinib and gefitinib, block ATP binding in the tyrosine kinase domain of EGFR (2). Phase II trials have demonstrated its safety, tolerability and antitumor activity in HNSCC patients (3-5). The low response rate of single agent erlotinib supports the use of combinations, which may augment its clinical utility and warrants investigation into biomarkers of response and their mechanism of action. Blood-based biomarkers are desirable due to the relative inaccessibility of tumor tissue for repeated sampling. Accurate and reproducible measurement of serum biomarkers may help elucidate disease pathogenesis and predict response to therapy. However clinical studies to date evaluating potential biomarkers have generally lacked placebo control groups or paired serum measurements.
Cross-communication between signaling systems is an essential aspect of the integration of extracellular stimuli into a limited number of pathways. In particular, G-protein-coupled receptors (GPCRs) activate the EGFR in HNSCC increasing proliferation and invasion. Several GPCR ligands are upregulated in HNSCC including prostaglandin E2 (PGE2) (6). Targeting of PGE2 using the COX-2 inhibitor celecoxib has been the most extensively studied in cancer patients. A Phase I trial of gefitinib plus celecoxib in HNSCC patients with recurrent/metastatic disease reported that this combination was well-tolerated and showed clinical activity (7).
Due to concerns about the adverse cardiovascular effects of COX-2 inhibitors less selective COX inhibitors are being evaluated in adjuvant and cancer prevention studies. Sulindac (an inhibitor of both COX-1 and COX-2) has been shown to inhibit the growth of HNSCC cells in vitro (6). The combination of celecoxib and gefitinib demonstrated synergistic growth inhibition in preclinical HNSCC models (6, 8).
Circulating biomarkers such as cytokines, chemokines and growth factors can be measured by a variety of assay formats including enzyme-linked immunosorbent assay, radioimmunoassay, bioassays and mass spectrometry (9, 10). Multiplex ELISA assays offer the theoretical advantage of requiring small sample volumes to measure a large number of analytes simultaneously. However, little data exist comparing the results between different multiplex technologies and single agent ELISA used on the same sample(s). In the context of a prospective double-blinded trial, we compared the effect of erlotinib with or without sulindac, to a placebo, in modulating serum biomarkers. We compared two commonly available technologies: multiplex sandwich ELISA (SearchLight™) and bead based assays (Luminex™). Using paired serum specimens from each patient, we investigated changes in the expression of a panel of putative biomarkers in three cohorts of patients who were treated either with erlotinib alone, erlotinib plus sulindac, or placebo.
Materials and Methods
Patient Selection
The clinical protocol was approved by the Institutional Review Board at the University of Pittsburgh, and registered at UPCI trial 05-045, IRB 0507063. Patients with histologically or cytologically confirmed HNSCC provided informed written consent before being enrolled into a biomarker study 1-2 weeks prior to surgical resection. Between December 2005 and December 2008, 41 HNSCC patients were randomly assigned and treated with erlotinib, erlotinib + sulindac or placebo at a 5:5:3 ratio, respectively, Serum was collected from a subset of patients who provided complete blood draws at all time points to investigate analyte alterations after neoadjuvant treatment (Table 1). Patients had no history of HNSCC, no prior therapy targeting the EGFR pathway, and no known severe hypersensitivity to sulindac or other non-steroidal anti-inflammatory drugs (NSAIDs), including aspirin. The patients were 18 or older, had clinical stage II, III or IVA disease without distant metastasis, as defined by the American Joint Committee on Cancer Staging System, Sixth edition and performance status 0 or 1. Sites of involvement included primary tumors of the oral cavity, oropharynx, hypopharynx, or larynx, but excluded primary tumors of the sinuses, paranasal sinuses, or nasopharynx, or unknown primary tumors.
Table 1.
Demographic Data for Patients with Serum Data Utilized in this Pilot Study
| Patient | Tx Assignment | Male/ Female |
Age at Diagnosis |
Location of Primary Tumor |
Tumor Stage |
|---|---|---|---|---|---|
| 1 | Erlotinib | Male | 57 | Hypopharynx | IVa |
| 2 | Erlotinib | Female | 65 | Oral Cavity | IVa |
| 3 | Erlotinib | Male | 59 | Oral Cavity | III |
| 4 | Erlotinib | Male | 68 | Oral Cavity | II |
| 5 | Erlotinib | Male | 66 | Oral Cavity | IVa |
| 9 | Erlotinib | Male | 67 | Larynx | III |
| 13 | Erlotinib | Male | 64 | Tonsil | II |
| 15 | Erlotinib | Female | 65 | Larynx | III |
| 16 | Erlotinib | Female | 61 | Oral Cavity | IVa |
| 23 | Erlotinib | Male | 46 | Oral Cavity | IVa |
| 6 | Erlotinib + Sulindac | Male | 58 | Oral Cavity | III |
| 7 | Erlotinib + Sulindac | Male | 59 | Oral Cavity | IVa |
| 8 | Erlotinib + Sulindac | Male | 53 | Oral Cavity | III |
| 14 | Erlotinib + Sulindac | Male | 56 | Oral Cavity | II |
| 17 | Erlotinib + Sulindac | Male | 64 | Oral Cavity | III |
| 20 | Erlotinib + Sulindac | Female | 57 | Oral Cavity | III |
| 21 | Erlotinib + Sulindac | Male | 51 | Oral Cavity | IVa |
| 22 | Erlotinib + Sulindac | Male | 64 | Oral Cavity | IV |
| 10 | Placebo | Male | 57 | Oral Cavity | IV |
| 11 | Placebo | Male | 33 | Oral Cavity | III |
| 12 | Placebo | Female | 75 | Oral cavity | III |
| 18 | Placebo | Male | 61 | Oral cavity | II |
| 19 | Placebo | Female | 46 | Oropharynx | II |
Patient Treatment
Subjects were randomized to a 7-14 day course of erlotinib (150 mg PO QD) plus sulindac (150 mg PO BID), erlotinib alone (150 mg PO QD) or placebo. The neoadjuvant therapy was discontinued one day prior to surgical resection. The patients, treating physicians and investigators were blinded to the treatment group status.
Collection and Storage of Blood Serum
Ten milliliters of peripheral blood were drawn from subjects using standard venipuncture techniques and allowed to clot. Blood was collected prior to neoadjuvant therapy (at the time of diagnostic biopsy) and on the day of surgery. Blood samples were collected without anticoagulant in red top vacutainers and allowed to coagulate for 15 to 30 minutes at room temperature. Sera were separated by centrifugation and all specimens were immediately aliquoted, frozen and stored in a dedicated −80°C freezer. No more than one freeze-thaw cycle was allowed for each sample.
Luminex™ Assays
The LabMAP technology (Luminex™) assays were performed in 96-well microplates in a format according to the protocol by Biosource International (Camarillo, CA). A filter-bottom, 96-well microplate was blocked for 10 minutes with bovine serum albumin. The differentially dyed polystyrene microspheres served as a solid support for the sandwich assay. A panel of serologic markers including cytokines, chemokines, growth and angiogenic factors and other markers was analyzed in the sera of patients. Detailed description of the analysis has been described in previous publications (9, 11, 12).The biomarkers analyzed in triplicate with the Luminex™ platform in this study were: CXCL5, G-CSF, IFNγ, IL-1α, IL-1β, IL-1ra, IL-6, CXCL8, IL-10, CCL2, CCL4, TPO, VEGF, GM-CSF, IP-10, CD40L, EGF, HGF, Eotaxin, Leptin, I-TAC
SearchLight™ Assays
The SearchLight™ technology (Thermo Scientific SearchLight Protein Array) utilizes a multiplexing sandwich-ELISA system based on chemiluminescent detection of analytes whose respective capture-antibodies are spotted in arrays within each well of a 96-well microplate. Calibrated protein standards and serum samples were added to coated wells in triplicate, incubated at room temperature, washed, and then biotinylated antibodies were added that specifically bind to the captured proteins. After incubation at room temperature and another round of washes, the wells were incubated with streptavidin-horseradish peroxidase conjugate. After another round of washes, the Super-Signal ELISA Femto Chemiluminescent Substrate was added. The plate was imaged using a CCD imaging system to capture the chemiluminescent signal from each spot in each well on the plate. The concentration of each analyte was quantified by comparing spot intensities for each unknown sample to the corresponding standard curves. To reduce interassay variations, all runs of the same multiplex assay were performed on the same day and each plate contained its own standard curve. The biomarkers analyzed with the SearchLight platform in this study were: TGF-α, IL-6, HGF and EGFR. These biomarkers were selected due to their biologic plausibility in contributing to GPCR-EGFR transactivation following optimization of antisera for multiplexing.
ELISA Assays
Single-analyte analyses of individual cytokines (HGF and IL-6 were performed in triplicate using R&D Systems kits (R&D Systems, Minneapolis, MN). Assays were performed according to the manufacturer’s instructions. All samples were run on a single plate to reduce assay variability.
Statistical analysis
Pre to post serum changes were calculated and tested for change with the signed rank test. This test was conducted for all patients irrespective of treatment assignment. To test whether pre to post change depended upon treatment group, a Kruskal-Wallis test was conducted. The false discovery rate was controlled for a large number of tests (Luminex™) by the method of Storey and Tibshirani (13). The smaller Searchlight™ series p values were adjusted by the method of Benjamini and Hochberg (14). A finding was reported as positive if the estimated false discovery rate (FDR) was < 10%. Concordance among different assays for the same serum samples was investigated by linear regression with transformations as needed for variance stabilization and linearity. We calculated an 80% power to detect a ‘difference’ in biomarker(s) if one group was 2.5 standard deviations greater than another in expression level of a particular analyte.
Results
Serum Analyte Changes Assessed by Luminex™ are not Treatment-Specific
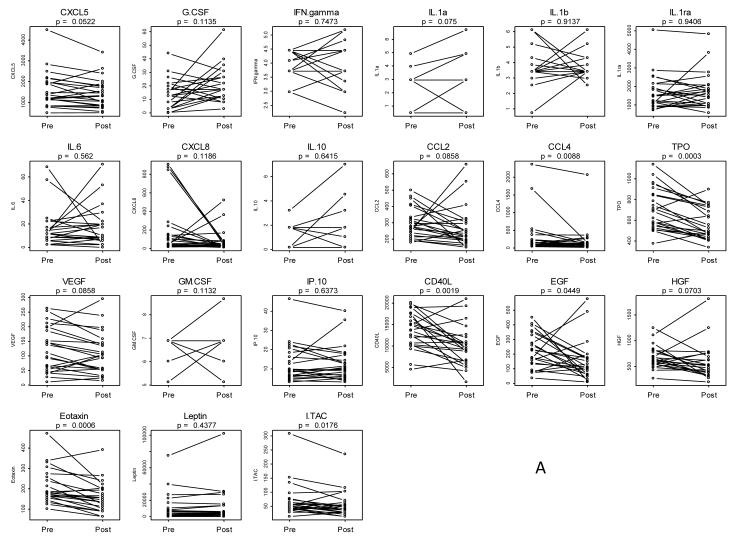
Paired pre-treatment and post-treatment sera were obtained from 23 patients for analysis with the Luminex™ platform. Table 1 shows the demographic and clinicopathologic characteristics of this cohort. Included in this cohort were patients treated only with erlotinib (n=10), patients treated with erlotinib plus sulindac (n=8) and patients who received placebo (n=5). We selected analytes based on a subset of a previously established panel of biomarkers using Luminex™ in HNSCC (9, 11, 15). These markers were selected for their association with one or more of the pathways modulated in this trial: EGFR/STAT3 (EGF, VEGF, EGFR, HGF, IL-6, IL-10): STAT1 ( IFN-γ, IL-1α, IL1β, IL1Rα, I-TAC, IP-10), and inflammatory pathways (CXCL5, CXCL8, Eotaxin, GM-CSF, CCL4). Aliquots of the same serum samples were analyzed by each assay to limit any variability in serum processing. Thirty one analytes were tested but eight were removed due to values below the detection liming missing data (50% to 100%) and two were removed for having at most four unique values, leaving 21 analytes for analysis. Figure 1a shows the paired pre-treatment and post-treatment levels for the 21 analytes assayed with the Luminex™ platform (CXCL5, G-CSF, IFNγ, IL-1α, IL-1β, IL-1ra, IL-6, CXCL8, IL-10, CCL2, CCL4, TPO, VEGF, GM-CSF, IP-10, CD40L, EGF, HGF, Eotaxin, Leptin, I-TAC). Five analytes were significantly changed (all were decreased) after limiting false discovery to 10%: TPO, Eotaxin, CD40L, CCL4 and ITAC. Table 2 summarizes pre to post changes in each analyte by assay.
Figure 1. Pre to post changes in 23 serum analytes measured by Luminex™.
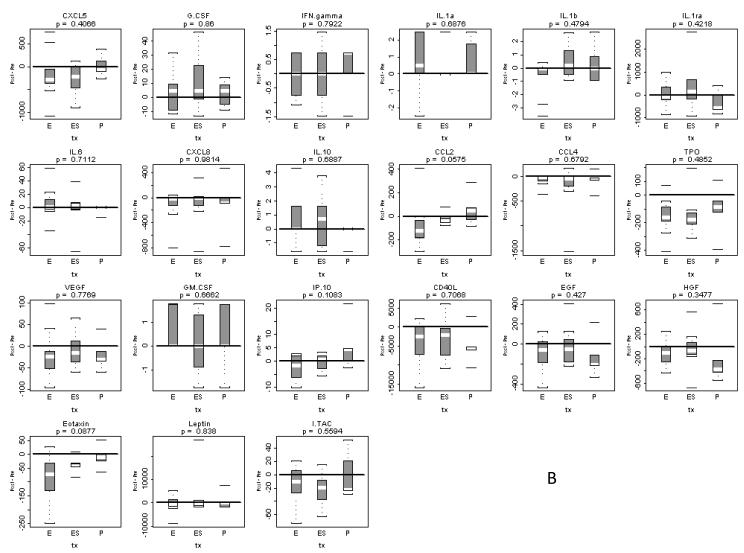
(A) Individual patients are depicted with a straight line connecting pre treatment to post treatment. The unadjusted p-value resulting from a two-tailed signed rank test is provided above each graph. Five analytes were significantly changed after limiting false discovery to 10%: TPO, Eotaxin, CD40L, CCL4 and ITAC. (B) Box and whisker plots of patient differences shown by treatment group. The randomly assigned groups are shown on the X axis, E = erlotinib, ES = erlotinib + sulindac, P = placebo. The p-value for the two-tailed Kruskal-Wallis test of group equality is shown above each plot.
Table 2.
Serum Analyte Changes by Platform
| Median Analyte Ratio and Difference Pre – Post Change |
||||||
|---|---|---|---|---|---|---|
| Serum Analyte | Luminex | Searchlight | ELISA | |||
| Ratio | Diff | Ratio | Diff | Ratio | Diff | |
| CXCL5 | .90 | −209 | --- | --- | --- | --- |
| GCSF | 1.89 | 4.64 | --- | --- | --- | --- |
| IFNγ | .77 | 0 | --- | --- | --- | --- |
| IL-1α | .83 | 0 | --- | --- | --- | --- |
| IL-1β | .77 | 0 | --- | --- | --- | --- |
| IL-1ra | 1.03 | −75.6 | --- | --- | --- | --- |
| IL-6 | 1.00 | 0 | 1.65 | 1.6 | 1.73 | 4.81 |
| CXCL8 | .81 | −16.8 | --- | --- | --- | --- |
| IL-10 | .62 | 0 | --- | --- | --- | --- |
| CCL2 | .84 | −42.3 | --- | --- | --- | --- |
| CCL4 | .74 | −24.8 | --- | --- | --- | --- |
| TPO | .79 | −152 | --- | --- | --- | --- |
| VEGF | .84 | −25.0 | --- | --- | --- | --- |
| GMCSF | .93 | 0 | --- | --- | --- | --- |
| IP10 | .93 | 0 | --- | --- | --- | --- |
| CD40L | .75 | −3038 | --- | --- | --- | --- |
| EGF | .66 | −70.6 | --- | --- | --- | --- |
| HGF | .82 | −100 | .90 | −85 | .74 | −320 |
| Eotaxin | .76 | −39.7 | --- | --- | --- | --- |
| Leptin | .88 | −544 | --- | --- | --- | --- |
| ITAC | .81 | −21.4 | --- | --- | --- | --- |
| TGF-α | --- | --- | .53 | −2.8 | --- | --- |
| EGFR | --- | --- | .92 | −3668 | --- | --- |
Bold denotes significant changes with FDR < 10%, NA=not applicable
To assess whether serum analyte changes were associated with treatment we plotted changes by treatment group and tested the differences between groups. Figure 1b illustrates box and whisker plots of paired differences (post-treatment – pre-treatment) by treatment group for the 21 analytes assayed with the Luminex™ protocol. Several analytes, most notably Eotaxin, TPO and CD40L, revealed that post-treatment analyte expression levels were significantly lower than pre-treatment levels in the erlotinib and erlotinib plus sulindac treatment groups. However, when changes in analyte expression were analyzed in the placebo, control changes were similar to the other treatment groups and, based on the Kruskal-Wallis test, there were no statistically significant differences between the three cohorts Therefore, the Luminex™ assay could detect serum analyte changes with therapy, but these changes appeared not to be treatment-specific. These findings raised the question whether Luminex™ was insufficiently sensitive to detect intergroup differences.
Serum Analyte Changes Assessed by SearchLight™ are not Treatment-Specific
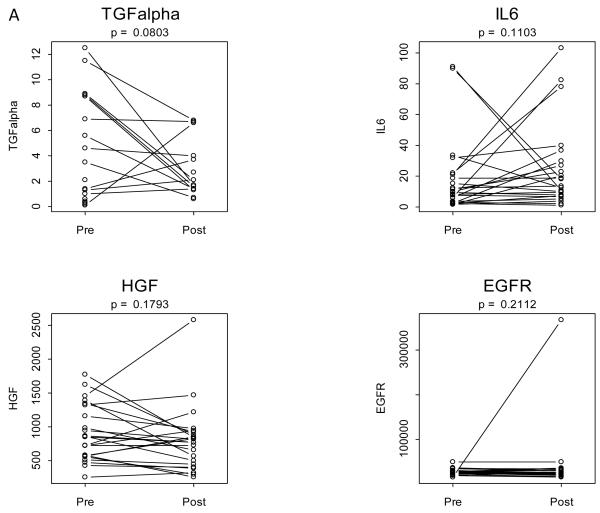
Using the same 23 patients’ sera analyzed by Luminex™, we used the SearchLight™ platform for comparison of intergroup analyte changes. We selected four analytes from the initial Luminex™ panel (TGF-α, IL-6, HGF and EGFR) based on previous reports of their role in GPCR-EGFR transactivation in HNSCC tumors and the biologic plausibility of their secretion into serum. In addition, several analytes have been reported to have prognostic significance in cancer patients, including HNSCC. Specifically, TGF-α has been shown to contribute to GPCR-EGFR transactivation in HNSCC preclinical models and prior studies have reported increased serum TGF-α following erlotinib treatment in lung cancer patients (16). Serum IL-6 and/or HGF levels have been reported to have prognostic significance in HNSCC (17, 18). Soluble EGFR has been proposed as a serum biomarker to monitor response to treatment in cancer patients (19). We designed and optimized multiplexed assays of these four analytes using the SearchLight™ platform (20). Figure 2a demonstrates the changes in pre-treatment and post-treatment expression of these four analytes. These data suggested that TGF-α may have decreased following treatment, although false discovery rate for TGF-α was 32%. Figure 2b illustrates box and whisker plots of the paired differences (post-treatment – pre-treatment) for the analytes assayed with the SearchLight™ protocol (TGF-α, IL-6, HGF and EGFR) presented by treatment group, and analyzed using the Kruskal-Wallis test of equality. Changes in analyte expression from pre-treatment to post-treatment levels did not identify a statistically different level of expression between the three treatment groups (Figure 2b, Table 2). Similar to the Luminex™ results, levels of expression between the three treatment groups did not differ significantly for any single analyte studied: TGF-α (p=0.3406), IL-6 (p=0.486), HGF (p=0.3334) and EGFR (p=0.8598). In order to compare serum with tumor EGFR levels, we also isolated 44 tumor - serum pairs from pre and post sampling. There was correlation found between these variables (data not shown).
Figure 2. Pre to post changes in 4 serum analytes measured with the SeachLight platform.
(A) Individual patients are depicted with a straight line connecting pre treatment to post treatment. The p-value resulting from a two-tailed signed rank test is provided above each graph. (B) Box and whisker plots of patient differences shown by treatment group. The randomly assigned groups are shown on the x asis, E = erlotinib, ES = erlotinib + sulindac, P = placebo. The p-value for the two-tailed Kruskal-Wallis test of group equality is shown above each plot.
ELISA Assays of HGF and IL-6 Reveal Effects That Were Undetected With Multiplex Systems
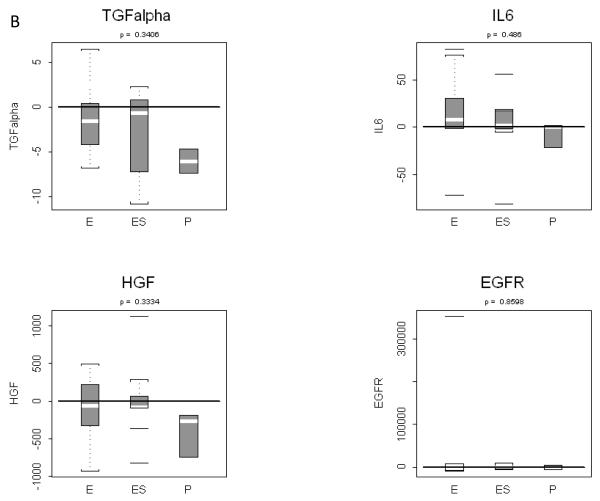
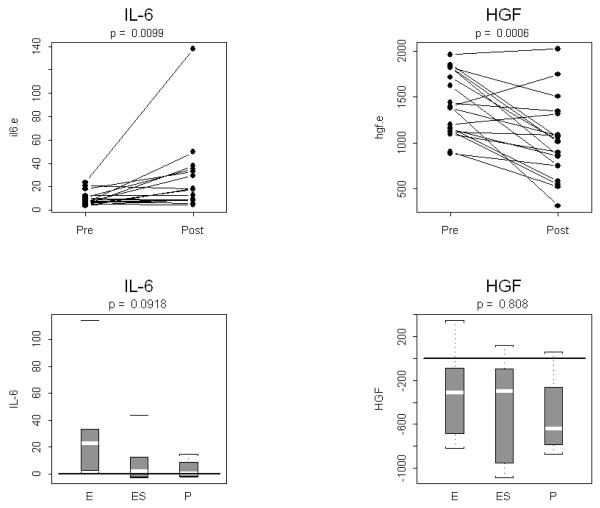
Single ELISA assays were conducted for IL-6 and HGF. Assay results were used to compute pre to post treatment changes and to assess treatment specificity, analogous to the outcome of multiplex assay results. We found a strong and significant increase in HGF and strong decrease in HGF (see Table 2 and Figure 3). Neither change was detectable using either of the two multiplex assays described above. However, consistent with multiplex results for these two analytes, no treatment specific effect was found.
Figure 3. Pre to post changes in 4 serum analytes measured with the ELISA assay.
(A) Individual patients are depicted with a straight line connecting pre treatment to post treatment. The p-value resulting from a two-tailed signed rank test is provided above each graph. (B) Box and whisker plots of patient differences shown by treatment group. The randomly assigned groups are shown on the x-axis, E = erlotinib, ES = erlotinib + sulindac, P = placebo. The p-value for the two-tailed Kruskal-Wallis test of group equality is shown above each plot.
Comparison of Luminex™, Searchlight™ ELISA assays for IL-6 and HGF
Multiplexed assays are known to vary in the precise quantification of analyte expression as compared to the gold standard of a single analyte ELISA. In order to compare the results from the multiplex assays, HGF and Il-6 expression were assayed utilizing an individual single analyte ELISA quantification, in addition to measurement with Luminex™ and SearchLight™ since these were the only two analytes included in both multiplex platforms.
The ELISA experiments measuring the expression of HGF in the serum samples demonstrated excellent reproducibility with a coefficient of variation of 9.2% and an intraclass correlation coefficient of 0.945. IL-6 also had excellent reproducibility with a coefficient of variation of 10.9% and an intraclass correlation coefficient of .993.
Biomarker expression levels differ across all three platforms
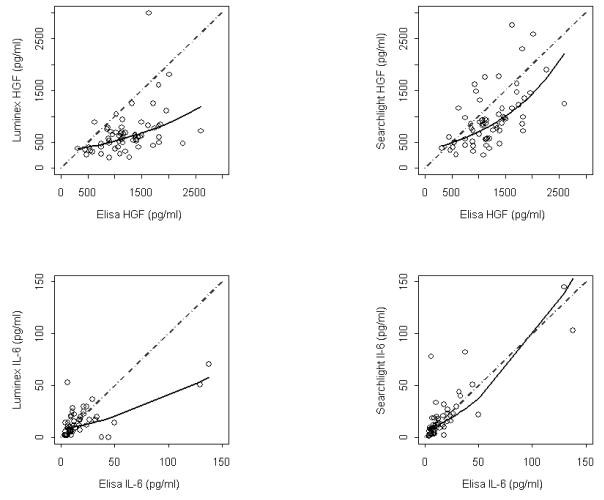
With the outcome of two serum analytes (IL-6 and HGF) available from all three platforms we sought to compare the three analytical methods using the same specimens (Table 2). We selected ELISA as the reference standard to compare with Luminex™ and Searchlight™ results that were common to the three methods. Figure 4 shows the agreement between Luminex™ (left column) and Searchlight™ (right column). Linear regression models were fit to the data points and the fitted values are shown as a solid line. This line can be visually compared to the diagonal dashed line showing hypothetical perfect agreement. Substantial underestimation by Luminex™ measurement of HGF and IL-6 is evident. Greater agreement was observed between ELISA and Searchlight™ results. A scatterplot matrix (data not shown) of common measurements revealed that as compared to ELISA, both SearchLight™ and Luminex™ appear to have a restricted dynamic upper range. The constriction within the upper range of expression levels suggests that cross-reactivity and antagonism due to interactions between antibodies may dampen the dynamic range of measurements pertaining to a single analyte in a multiplex format.
Figure 4. Concordance of assay platforms.
Sixty-six patient serum samples of two analytes IL-6 and HGF were measured by ELISA, Luminex and Searchlight. ELISA was regarded as the reference standard and plotted on the X axis of all plots. A linear regression model was fit for each pair with Luminex (left column) or Searchlight (right column) serving as the dependent variable. Based on examination of residuals either log or square root transformations were required for selection of correct models. The solid lines are predicted values from the best fitting regression models. A dashed diagonal reference has been added to show hypothetical perfect agreement. When compared to ELISA, the Luminex assay has a restricted linear range and consistently underestimates analyte expression.
Discussion
The discovery of novel molecular therapeutic agents has led to new opportunities in patient treatment. Understanding the pathways that are modulated by these agents may play a vital role in harnessing the mechanisms by which these drugs work, and to potentially improve patient selection. In the present study, patients with resectable HNSCC were randomized to treatment for up to two weeks with erlotinib, erlotinib with sulindac, or with placebo alone, in the neoadjuvant setting. The analysis of pre-treatment and post-treatment sera permitted the measurement of alterations in potential biomarkers in treated patients.
Through a novel neo-adjuvant study design and inclusion of the placebo group, this study provides a unique opportunity to compare alterations in putative serum biomarkers with two pharmaceutical drugs known to inhibit tumor growth in preclinical model systems. A placebo control was utilized in order to investigate the specificity of group differences. Several of the selected biomarkers revealed increased or decreased post-treatment expression for patients treated either with erlotinib alone or with erlotinib plus sulindac (Figure 1b). In most instances, these changes were in the same direction and to the same degree in both groups. In the absence of a placebo control group these changes could have been incorrectly viewed as positive or negative indicators of drug treatment. However, similar effects observed in the placebo treated cohort indicate that the changes were likely intrinsic to the disease process itself and/or assay variability as opposed to the drug regimen. Possible sources for additional alterations seen in the post-treatment serum across each treatment group may indicate that such analytes are markers of a stress response in the peri-operative setting that could be due to fasting, anesthesia and/or other perioperative parameters.
Limitations of this study include the small sample size (n=23), incomplete sampling of patients enrolled on the clinical trial (n = 41 patients treated) and the number of analytes studied. However we note that the sample size is sufficient to detect large effect sizes, a few of which could reasonably be expected if the serum biomarkers were truly indicative of treatment effects. For example, we would have 80% power to detect a treatment – related difference in biomarker(s) if one group was 2.5 standard deviations greater than another in level of expression in a particular analyte. Exploratory studies often consider hundreds or thousands of analytes although false discovery can occur with even the 21 analytes evaluated here. These restrictions may have impacted the ability to find statistically significant changes in analytes in this study population. In addition, the one to two week treatment period was not considered therapeutic and thus we had no clinical endpoint with which to correlate the molecular changes with prognostic clinical information.
Since the discovery and development of EGFR TKIs there has been a rapid expansion in the number of studies investigating clinical efficacy of these agents for the treatment of various types of cancer. However, only a limited number of studies have sought to assess alterations in biomarkers induced by the administration of such agents in HNSCC. In addition, the majority of identified biomarkers derived from cell lines or baseline patient tissue. Hickinson et al. found higher mRNA expression levels of E-cadherin, TGF-alpha, amphiregulin, PAK6, and EGFR for gefitinib-sensitive HNSCC lines (21). Pernas et al. identified that gefitinib sensitivity correlated with p-AKT and p-STAT3 activation in cell lines (22). Agulnik and coworkers reported that elevated baseline tumor levels of p27 and p-STAT3 predicted prolonged time to progression and overall survival and noted that with erlotinib treatment a decrease in p-EGFR, p-NFkB and p27 correlated with increased time to progression and overall survival (23). Others have reported that erlotinib treatment was associated with decreased p-EGFR expression and p27 upregulation in post-treatment specimens (24). Another group showed a decrease in phosphorylated tyrosine residues and phosphorylated ERK immunostaining in tumors from patients with locally advanced nonmetastatic HNSCC treated with erlotinib (25).
In terms of serum biomarkers, Chung et al., utilized serum mass spectrometry in patients treated with cetuximab, EGFR-TKIs or chemotherapy for recurrent or metastatic HNSCC to identify possible biomarkers in patient serum samples (10). They utilized a MALDI MS data analysis algorithm previously used for non-small cell lung cancer to predict overall survival benefit in those patients treated with gefitinib and erlotinib/bevacizumab using a set of predefined mass-to-charge features. However, the identity of the peptides constituting the MS signature in this study is currently unknown.
Prior studies focused on a small number of putative biomarkers and did not offer the opportunity for analysis of a large number of biomarkers via multiplexing capabilities afforded by the technologies utilized here. One group compared FAST Quant, SearchLight and Luminex platforms in human samples (26). In paired comparisons there were a number of statistically significant differences for individual cytokines, but these varied between platforms, suggesting that subtle differences may be due to the use of different sources for antibody pairs and standards.
Multiplex assays offer the potential for a large amount of data from small samples. Since different techniques may yield different numerical results, a single assay technique should be used to complete each project to generate optimum consistency. Toedtet et al. found that the solid-phase multiplex assays can perform reliably within a given platform (27). However, they strongly recommended testing the possible combinations of antibodies on the array in order to reduce the incidence of cross-reaction and enable combination of analytes that require similar serum dilutions to be placed upon the same array. Khan et al. compared multiplex bead arrays and found that cytokine levels follow similar patterns across different platforms (28), but the absolute levels of cytokines measured varied by manufacturer. Accordingly, they suggested that the quantitative values from within a specific platform were reproducible, but there was too much variation to allow comparison of results between different platforms.
Differences between platforms can be due to numerous sources, including matrix effects, antibody cross-reactivity with other cytokines, the accuracy of the standardization of the calibration curve, the specific antibodies used for capture and detection, the use of serum or plasma for sample collection, and proper buffering of the sample to minimize matrix interference with the detection of the analytes. Potential reasons for variability in the output generated from these assays also include random errors of the test instrument, such as bead heterogeneity and carry-over of beads from previous wells as well as the presence of interfering substances in blood. Furthermore, the absolute value of analytes measured by different platforms often differs due to the utilization of different antibodies for the recognition of analytes. There are very few reports on use of multiplex arrays to evaluate pharmacodynamic responses to targeted therapy in early phase clinical trials. Backen et al. used two multiplex SearchLight ELISA platform arrays to evaluate potential angiogenesis biomarkers in serum from eight ovarian cancer patients who were not receiving therapy (29). They found no evidence that the multiplex signals from one analyte impinged on another or that antibody cross-reactivity occurred. Allen et al. prospectively studied 30 patients with oropharyngeal SCC who were receiving chemoradiation, comparing cytokine levels at baseline to serial measurements at three month intervals. They determined cytokine concentrations in serum using a Luminex™ multiplex assay and found that increases in serum levels for individual factors IL6, IL8, VEGF, HGF and GRO-1 were significantly associated with decreased cause-specific survival up to 12 months post-treatment. However, this study also included only a small number of patients (n=30) and in the absence of a placebo control group, the specificity of the findings related to treatment cannot be evaluated.
The results of the present study provides further insights into issues that need to be addressed to decide which multiplex platform to utilize studying the context of a clinical trial. Each platform differs in the sample volume required for the assay. SearchLight™ and Luminex™ offer the capability to greatly reduce the sample volume required compared to individual ELISA experiments. This can be an important factor when sample volume available is limited. However, in the current SearchLight™ analysis described in this paper although only four biomarkers were studied, two separate arrays were required due to cross-reactivity in the analytes selected for review, thus only doubling the efficiency as compared to single ELISAs. Therefore, the advantages of custom arrays may be mitigated by difficulties with cross-compatibility that may limit the number of analytes that can be assayed in a single experiment. Another important issue relates to the specificity of the analyte quantification. Our data revealed constriction within the upper range of expression levels in experiments with SearchLight™ and Luminex™ platforms. These results suggest that cross-reactivity and antagonism due to interactions between antibodies may alter the results of a single analyte in a multiplex format. Comparison of the results from the three platforms indicated potential differences due to precision and bias. The linear regression of the data from paired analyses from the platforms differed from estimation of potential ideal agreement. In the absence of bias the regression line would overlay a line of perfect agreement. Comparison of SearchLight™ and Luminex™ data for IL-6 and HGF to ELISAs revealed modest bias for the Searchlight™ assay and considerable bias for the Luminex™ assay (Figure 4). Using the more reliable ELISA assay we report significant changes in IL-6 or HGF that would remain undetected had we been limited to either multiplex assay. We believe the attenuated dynamic range and underestimation of IL-6 and HGF by Luminex™ could compromise the ability to detect predictive or prognostic use of these biomarkers. The present study highlights the value of incorporating placebo controls into biomarker studies and clinical trials, when ethically justified. A placebo group is necessary to control for group effects and permits the opportunity to assess the pharmacodynamic impact of molecular targeting agents, such as EGFR TKIs, in serum. Furthermore, although this report illustrates the ability of multiplex ELISA platforms to provide more information about a wider panel of analytes using less biological material, this additional data may come at the expense of reflecting the true dynamic range of expression of each individual analyte tested.
Acknowledgments
Grant Support – P50 CA097190, P30 CA047904, R01 DE19727
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Argiris A, Karamouzis MV, Raben D, Ferris RL. Head and neck cancer. Lancet. 2008;371(9625):1695–709. doi: 10.1016/S0140-6736(08)60728-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moyer JD, Barbacci EG, Iwata KK, Arnold L, Boman B, Cunningham A, et al. Induction of apoptosis and cell cycle arrest by CP-358,774, an inhibitor of epidermal growth factor receptor tyrosine kinase. Cancer Res. 1997;57(21):4838–48. [PubMed] [Google Scholar]
- 3.Perez-Soler R, Chachoua A, Hammond LA, Rowinsky EK, Huberman M, Karp D, et al. Determinants of tumor response and survival with erlotinib in patients with non--small-cell lung cancer. J Clin Oncol. 2004;22(16):3238–47. doi: 10.1200/JCO.2004.11.057. [DOI] [PubMed] [Google Scholar]
- 4.Gordon AN, Finkler N, Edwards RP, Garcia AA, Crozier M, Irwin DH, et al. Efficacy and safety of erlotinib HCl, an epidermal growth factor receptor (HER1/EGFR) tyrosine kinase inhibitor, in patients with advanced ovarian carcinoma: results from a phase II multicenter study. Int J Gynecol Cancer. 2005;15(5):785–92. doi: 10.1111/j.1525-1438.2005.00137.x. [DOI] [PubMed] [Google Scholar]
- 5.Soulieres D, Senzer NN, Vokes EE, Hidalgo M, Agarwala SS, Siu U. Multicenter phase II study of erlotinib, an oral epidermal growth factor receptor tyrosine kinase inhibitor, in patients with recurrent or metastatic squamous cell cancer of the head and neck. J Clin Oncol. 2004;22(1):77–85. doi: 10.1200/JCO.2004.06.075. [DOI] [PubMed] [Google Scholar]
- 6.Thomas SM, Bhola NE, Zhang Q, Contrucci SC, Wentzel AL, Frelino ML, et al. Cross-talk between G protein-coupled receptor and epidermal growth factor receptor signaling pathways contributes to growth and invasion of head and neck squamous cell carcinoma. Cancer Res. 2006;66(24):11831–39. doi: 10.1158/0008-5472.CAN-06-2876. [DOI] [PubMed] [Google Scholar]
- 7.Wirth LJ, Haddad RI, Lindeman NI, Zhao X, Lee JC, Joshi VA, et al. Phase I study of gefitinib plus celecoxib in recurrent or metastatic squamous cell carcinoma of the head and neck. J Clin Oncol. 2005;23(28):6976–81. doi: 10.1200/JCO.2005.02.4182. [DOI] [PubMed] [Google Scholar]
- 8.Chen Z, Zhang X, Li M, Wang Z, Wieand HS, Grandis JR, et al. Simultaneously targeting epidermal growth factor receptor tyrosine kinase and cyclooxygenase-2, an efficient approach to inhibition of squamous cell carcinoma of the head and neck. Clin Cancer Res. 2004;10(17):5930–9. doi: 10.1158/1078-0432.CCR-03-0677. [DOI] [PubMed] [Google Scholar]
- 9.Linkov F, Lisovich A, Yurkovetsky Z, Marrangoni A, Velikokhatnaya L, Nolen B, et al. Early detection of head and neck cancer: development of a novel screening tool using multiplexed immunobead-based biomarker profiling. Cancer Epidemiol Biomarkers Prev. 2007;16(1):102–7. doi: 10.1158/1055-9965.EPI-06-0602. [DOI] [PubMed] [Google Scholar]
- 10.Chung CH, Seeley EH, Roder H, Grigorieva J, Tsypin M, Roder J, et al. Detection of tumor epidermal growth factor receptor pathway dependence by serum mass spectrometry in cancer patients. Cancer Epidemiol Biomarkers Prev. 2010;19(2):358–65. doi: 10.1158/1055-9965.EPI-09-0937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hathaway B, Landsittel DP, Gooding W, Whiteside TL, Grandis JR, Siegfried J, et al. Multiplexed analysis of serum cytokines as biomarkers in squamous cell carcinoma of the head and neck patients. Laryngoscope. 2005;115(3):522–7. doi: 10.1097/01.mlg.0000157850.16649.b8. [DOI] [PubMed] [Google Scholar]
- 12.Linkov F, Ferris RL, Yurkovetsky Z, Marrangoni A, Velikokhatnaya L, Gooding W, et al. Multiplex analysis of cytokines as biomarkers that differentiate benign and malignant thyroid diseases. Proteomics Clin Appl. 2008;2(12):1575–85. doi: 10.1002/prca.200780095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Storey J, Tibshirani R. Statistical significance of genomewide studies. Proc Natl Acad Sci (2003) U S A. Aug 5;100(16):9440–5. doi: 10.1073/pnas.1530509100. Epub 2003 Jul 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9(7):811–8. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 15.Wang J, Seethala RR, Zhang Q, Gooding W, van Waes C, Hasegawa H, et al. Autocrine and paracrine chemokine receptor 7 activation in head and neck cancer: implications for therapy. J Natl Cancer Inst. 2008;100(7):502–12. doi: 10.1093/jnci/djn059. [DOI] [PubMed] [Google Scholar]
- 16.Zhou S, Ren S, Yan L, Zhang L, Tang L, et al. Clinical efficacy of erlotinib in patients previously treated for advanced non-small cell lung cancer. Respirology. 2009;14(5):709–15. doi: 10.1111/j.1440-1843.2009.01564.x. [DOI] [PubMed] [Google Scholar]
- 17.Meyer F, Samson E, Douville P, Duchesne T, Liu G. Serum prognostic markers in head and neck cancer. Clin Cancer Res. 2010;16(3):1008–1015. doi: 10.1158/1078-0432.CCR-09-2014. [DOI] [PubMed] [Google Scholar]
- 18.Allen C, Duffy S, Teknos T, Islam M, Chen Z, Albert PS, et al. Nuclear Factor-{kappa}B-Related Serum Factors as Longitudinal Biomarkers of Response and Survival in Advanced Oropharyngeal Carcinoma. Clin Cancer Res. 2007;13(11):3182–90. doi: 10.1158/1078-0432.CCR-06-3047. [DOI] [PubMed] [Google Scholar]
- 19.Baron AT, Wilken JA, Haggstrom DE, Goodrich ST, Maihle NJ. Clinical implementation of soluble EGFR (sEGFR) as a theragnostic serum biomarker of breast, lung and ovarian cancer. IDrugs. 2009;12(5):302–8. [PubMed] [Google Scholar]
- 20.Egloff A, Feinstein TM, Joyce SC, Kelly LA, Panelli MC, Yang T, et al. Blood biomarker modulation with dasatinib and cetuximab in patients with advanced solid malignancies. J Clin Oncol. 2009;27(15Suppl) abstr 6034. [Google Scholar]
- 21.Hickinson DM, Marshall GB, Beran GJ, Varella-Garcia M, Mills EA, South MC, et al. Identification of biomarkers in human head and neck tumor cell lines that predict for in vitro sensitivity to gefitinib. Clin Transl Sci. 2009;2(3):183–92. doi: 10.1111/j.1752-8062.2009.00099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pernas FG, Allen CT, Winters ME, Yan B, Friedman J, Dabir B, et al. Proteomic signatures of epidermal growth factor receptor and survival signal pathways correspond to gefitinib sensitivity in head and neck cancer. Clin Cancer Res. 2009;15(7):2361–72. doi: 10.1158/1078-0432.CCR-08-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Agulnik M, da Cunha Santos G, Hedley D, Nicklee T, Dos Reis PP, Ho J, et al. Predictive and pharmacodynamic biomarker studies in tumor and skin tissue samples of patients with recurrent or metastatic squamous cell carcinoma of the head and neck treated with erlotinib. J Clin Oncol. 2007;25(6):2184–90. doi: 10.1200/JCO.2006.07.6554. [DOI] [PubMed] [Google Scholar]
- 24.Calvo E, Malik SN, Siu LL, Baillargeon GM, Irish J, Chin SF, et al. Assessment of erlotinib pharmacodynamics in tumors and skin of patients with head and neck cancer. Ann Oncol. 2007;18(4):761–7. doi: 10.1093/annonc/mdl495. [DOI] [PubMed] [Google Scholar]
- 25.Thomas F, Rochaix P, Benlyazid A, Sarini J, Rives M, Lefebvre JL, et al. Pilot study of neoadjuvant treatment with erlotinib in nonmetastatic head and neck squamous cell carcinoma. Clin Cancer Res. 2007;13(23):7086–92. doi: 10.1158/1078-0432.CCR-07-1370. [DOI] [PubMed] [Google Scholar]
- 26.Lash GE, Scaife PJ, Innes BA, Otun HA, Robson SC, Searle RF, et al. Comparison of three multiplex cytokine analysis systems: Luminex, SearchLight and FAST Quant. J Immunol Methods. 2006;309(1-2):205–8. doi: 10.1016/j.jim.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 27.Toedter G, Hayden K, Wagner C, Brodmerkel C. Simultaneous detection of eight analytes in human serum by two commercially available platforms for multiplex cytokine analysis. Clin Vaccine Immunol. 2008;15(1):42–8. doi: 10.1128/CVI.00211-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khan IH, Krishnan VV, Ziman M, Janatpour K, Wun T, Luciw PA, et al. A comparison of multiplex suspension array large-panel kits for profiling cytokines and chemokines in rheumatoid arthritis patients. Cytometry B Clin Cytom. 2009;76(3):159–68. doi: 10.1002/cyto.b.20452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Backen AC, Cummings J, Mitchell C, Jayson G, Ward TH, Dive C. ‘Fit-for-purpose’ validation of SearchLight multiplex ELISAs of angiogenesis for clinical trial use. J Immunol Methods. 2009;342(1-2):106–14. doi: 10.1016/j.jim.2009.01.003. [DOI] [PubMed] [Google Scholar]