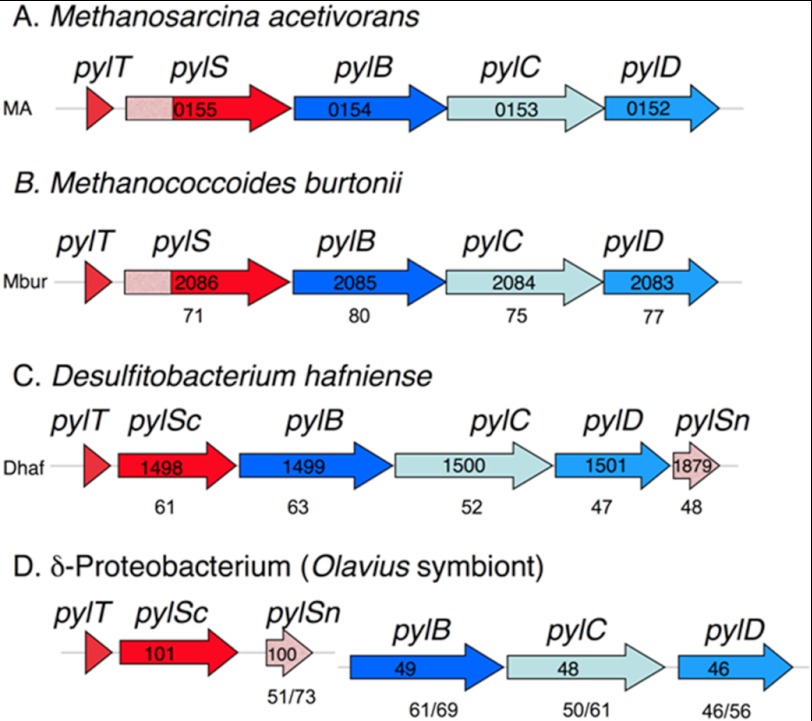
FIGURE 1.
Representative microbes with sequenced genomes that contain the pyl genes. A and B, the archaeal genomes are deposited at NCBI under accession numbers NC_003552 (A) and NC_007955 (B). C and D, the bacterial genomes have accession numbers NC_011830 (C) and AASZ01001259 as well as AASZ01001367 (D). The proteobacterial example is a gut symbiont sequenced in a metagenomic study of a marine worm. In all examples but D, the pyl genes are found in an apparent single transcriptional unit. In D, the pylTScSn genes and the pylBCD genes are found on separate contigs (groups of overlapping clones). The number labeling each gene is the locus number in the annotated genome. The numbers below genes in A–C show the percentage of similarity for each gene product when aligned with the corresponding M. acetivorans pyl gene product, in D the percentage of similarity to the corresponding gene products in M. acetivorans is shown, followed by the percentage of similarity to D. hafniense pyl gene products. Genes involved in the genetic encoding of pyrrolysine are in red, and those in biosynthesis of pyrrolysine are in blue. The pylSn genes are shown in pink, which matches the shading of the homologous 5′ region of the archaeal pylS genes.