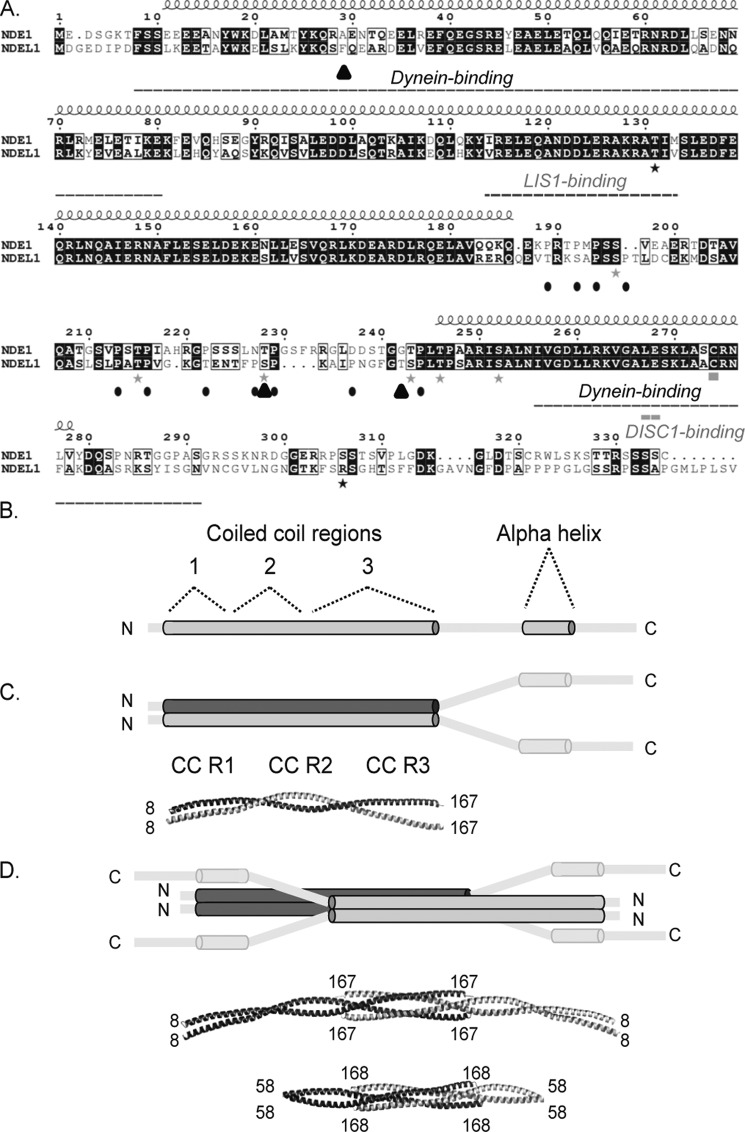
FIGURE 1.
Sequence and structure of NDE1 and NDEL1. A, sequence alignment between NDE1 and NDEL1. Indicated above the alignment is the consensus coiled-coil domain and predicted C-terminal α-helix for the two proteins (shown schematically in B). Phosphorylation sites for NDE1 and NDEL1 are shown as black and gray stars, respectively. NDE1 microcephaly frameshift mutations are shown as filled triangles. Binding sites described in text for dynein, DISC1, and LIS1 are indicated by dashed lines below alignment. Prolines lying between coiled-coil region and C-terminal α-helix are denoted by filled ovals. A palmytolation site is indicated by a filled rectangle. B, NDE1 and NDEL1 have an N-terminal coiled-coil domain (longer gray cylinder), which is made up of three coiled-coil regions (CC R): CC R1, CC R2, and CC R3. The two proteins are predicted to have a disordered C terminus (gray lines) containing a strongly predicted α-helix (shorter gray cylinder). The N and C termini are indicated. C, the NDEL1 N-terminal region was shown by crystallography (Protein Data Bank code 2V71) to dimerize via a parallel coiled-coil utilizing CC R1 (amino acids 9–44) and CC R2 (amino acids 44–99) (5). The structure of the C terminus remains unsolved. D, previously favored model of tetrameric assembly of full-length NDE1/NDEL1 (18) and, below, a structure determined by crystallography (the shorter tetramer fragment, consisting of amino acids 58–168 of Protein Data Bank code 2V66 and the longer tetramer fragment, consisting of amino acids 8–167 derived using symmetry operations from Protein Data Bank code 2V71). In the crystal structures, two NDEL1 dimers associate using CC R3 (amino acids 107–167) in an anti-parallel arrangement, to form tetramers (5).