Abstract
Ecologists hypothesize that community structure and stability affect productivity, sensitivity to invasion and extinction, and resilience and resistance to perturbations. Viewed in the context of the gut microbiome, the stability of the gut community is important for understanding the effects of antibiotics, diet change and other perturbations on host health and colonization resistance. Here we describe the dynamics of a self-contained community, the murine gut microbiome. Using 16S rRNA gene sequencing of fecal samples collected daily from individual mice, we characterized the community membership and structure to determine whether there were significant changes in the gut community during the first year of life. Based on analysis of molecular variance, we observed two community states. The first was observed in the 10 days following weaning and the second was observed by 15 days following weaning. Interestingly, these two states had the same bacterial populations, but those populations had different relative abundances in the two states. By calculating the root mean squared distances between samples collected in the early and late states for each mouse, we observed that the late state was more stable than the early state. This increase in stability was not correlated with increased taxonomic richness, taxonomic diversity, or phylogenetic diversity. In the absence of an experimentally induced perturbation, the second community state was relatively constant through 364 days post weaning. These results suggest a high degree of stability in the microbiome once the community reached the second state.
Keywords: 16S rRNA, 454 sequencing, communities, constancy, development, microbial ecology
Introduction
There has been recent interest in understanding how microbial community structures relate to the health of symbiotic, natural and engineered ecosystems.1-3 The dysbiosis hypothesis suggests that changes in human health are due to alterations in the structure of the microbial communities residing in that environment.4 The recent observation that enterotypes or community structures types that are independent of ethnicity, sex, and nationality, but are rather structured by an individual’s long-term diet emphasizes the point that changes in community structure do not necessarily have a negative impact on health or community productivity.5,6 Differences in community structure that do not result in a change in ecosystem health have been observed in non-host-associated environments as well.7 Taken together, a model emerges of a landscape where there are multiple community structure states where a community can reside. We hypothesize that these states vary in their resistance to perturbations, sensitivity to challenge by an exogenous population, and ability to fill the environment’s niche space. Ultimately, whether a state is “healthy” depends on all of these factors as well as the response of the specific environment to that state.
The mechanisms that drive change between these states are poorly understood. In host-associated communities, mechanisms that are likely to affect the structure of the microbiome include host genetics, alterations in the host immune system, ingestion of antimicrobials and diet.4 Differences in community stability are often associated with higher rates of extinction and colonization and these events can shift the overall community structure between healthy and unhealthy states.8 Understanding the stability of those states and how variation in stability affects variation in health can only be accomplished by characterizing the temporal dynamics of microbial communities. The importance of diversity and stability is a complex and contentious issue that has yet to be settled for plant, animal, or microbial communities.8 The lack of a clear correlation between diversity and stability emphasizes the need to develop a mechanistic understanding of the forces that stabilize microbial communities.
Recent studies have characterized the normal gut microbiota at multiple time points from the same person over long periods of time. Koenig and colleagues sampled an infant over 2.5 y and observed non-random successional patterns including an increase in phylogenetic diversity over the duration of the study.9 Caporaso and colleagues tracked the microbiota of two adults at four body sites for up to 15 mo and observed that few species were found at all time points, but many were found to be transient across the time courses.10 This suggests that the gut communities may be sampling species from a broader meta-community in a stochastic manner. Studies employing intensive sampling designs such as these indicate that the gut microbial community is more dynamic than suggested by studies that sample at two to four time points.1,11 These studies of the dynamics of “typical” microbiomes serve as a useful contrast to several studies that have used designs where the individual served as their own control to investigate the effects of diet change, antibiotic treatments, and the stability of enterotypes.6,12,13
While there is growing interest in performing intensive longitudinal sampling of humans, there has been less of a focus on studying the long-term dynamics of the microbial communities associated with experimentally tractable model animals.14 Most longitudinal studies either use a pre- and post-treatment sampling scheme or euthanize cohorts of animals at different time points to reconstruct a time course.15-18 This creates difficulties in interpreting the results of studies investigating models of pathogenesis where it is not possible to differentiate between the initial microbiome of those animals that succumb to disease and those that do not. Furthermore, with only pre- and post-treatment time points, it is not possible to ascertain whether the post-treatment sample represented a stable condition or whether the community was still experiencing perturbations imposed by the treatment.18 As an experimental system to study ecology, host-associated communities are advantageous because it is possible to sample them without perturbing the community and to make controlled manipulations of the host’s diet, environment, genetics, and initial microbiota. For example, several studies have used humanized and gnotobiotic mouse models to demonstrate a clear effect of diet changes on the structure of the murine microbiome.14,19,20
In this study, we investigated the dynamics of the gut microbiota obtained from 12 conventionally raised and colonized inbred mice living in a specific-pathogen free environment. We sought to test the hypothesis that the gut microbiome would be different in an immature mouse than in an adult. Our results supported this hypothesis and also demonstrated that as the animal matures their gut community became more stable.
Results
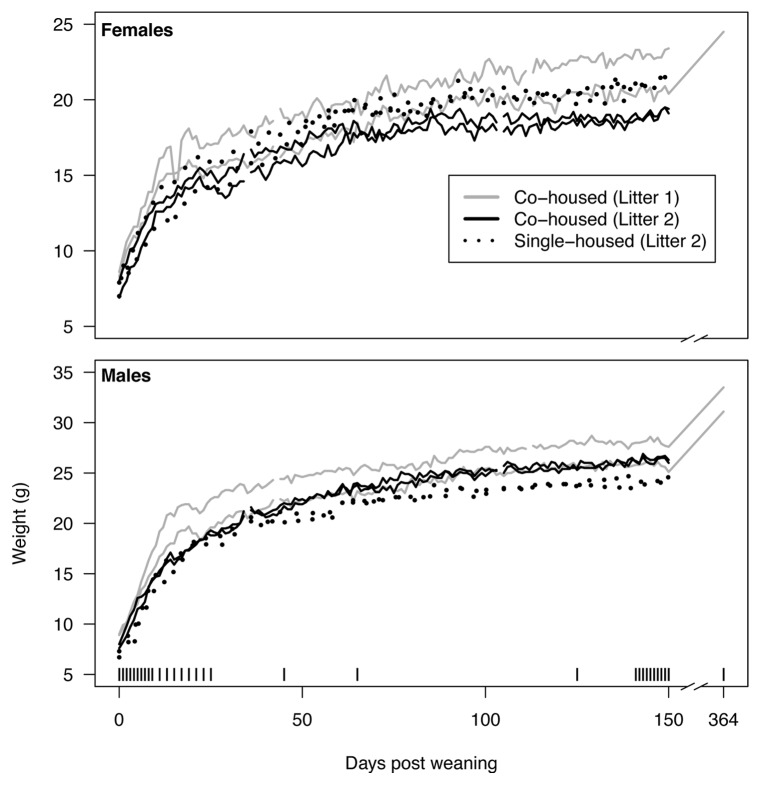
We obtained two litters of C57BL/6 inbred mice from a breeding colony at the University of Michigan. The first litter included 2 males and 2 females and the second litter included 4 males and 4 females. The two litters were born to different mothers 8 d apart. At 21 d after birth the mice from the first litter were weaned and separated by sex into two cages so that the 2 males and 2 females were each co-housed. In the second litter, 2 males and 2 females were also separated by sex into two cages. The remaining 4 mice from the second litter were also weaned and placed into four separate cages at 21 d after birth. At weaning weights and fresh feces were collected from each animal daily; this continued for the rest of the experiment. Immediately after weaning, all animals experienced rapid weight gain until approximately 25 d post weaning (dpw) when their growth rate slowed but continued to increase for the remainder of the experiment (Fig. 1). During the 150 dpw, the weight of the average female increased by 1.7-fold and the average male increased 2.3-fold. Among the three animals that were sampled until 364 dpw, the female gained an additional 20% and the two males gained 23%. Among the mice in the second litter, the median ratio of pairwise differences in weight between co-housed and single-housed animals was 0.86 for the females and 1.00 for the males. Co-housed males gained 1.9 g more than their single-housed littermates; however the co-housed females gained 1.9 g less than their single-housed littermates. Although co-housed mice from the first litter gained more weight and had a greater difference in weight than the co-housed mice from the second litter, Spearman correlation coefficients between weights of animals from the same litter were within the range of coefficients calculated between animals of different litters. Taken together, these results indicate that after controlling for age, genetics, diet, environment, sex, litter, and housing there was still a significant amount of variation in the weights of the mice from these litters.
Figure 1. Weights of mice over the first 364 d post weaning (dpw). For each litter two male mice were co-housed and two female mice were co-housed. In addition, two males and two females from the second litter were each individually housed. The vertical bars along the bottom indicate samples that were used for the microbiome analysis described in the current study.
To determine whether there was a change in the gut microbiome in the immature and mature animals, we used 454 FLX Titanium to sequence the V35 region of the bacterial 16S rRNA gene. We focused our microbial community analysis on the feces sampled at 0–9 dpw (“early”) and 141–150 dpw (“late”). Sequences were assigned to operational taxonomic units (OTUs) where the average distance between sequences was not greater than 0.03 and used to recreate their phylogeny. There were no statistically significant effects of sampling period (early vs. late) or sex on α diversity measures including the observed community richness (Sobs = 98.3 OTUs), inverse Simpson diversity index (1/D = 16.4 OTUs), Shannon diversity index (H’ = 3.26), and the phylogenetic diversity (PD = 5.54). We characterized the structure of the microbiome by calculating the pairwise distance between community structures using θYC (Fig. 2). Non-metric dimensional scaling plots demonstrated that the community structures of all 12 mice could be partitioned between the early and late time periods (AMOVA; all p < 10−4) and that there was not a significant effect of litter, single vs. co-housing, or sex on community structure. Similar results were observed when we used the weighted or unweighted UniFrac metrics (data not shown).
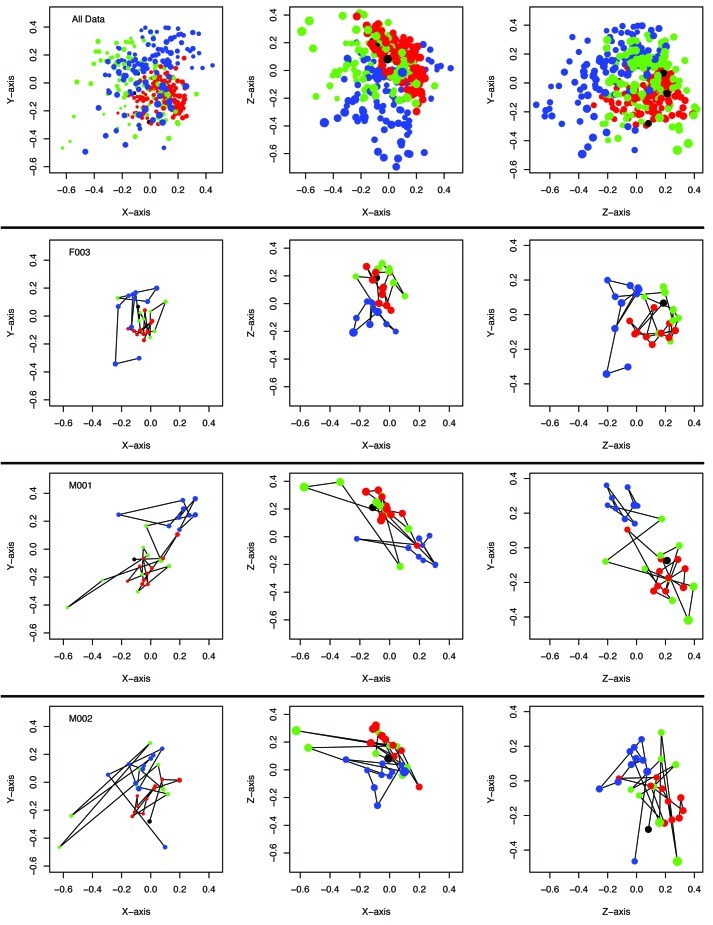
Figure 2. Temporal variation of the community structure of the murine gut microbiome for all 12 mice together as well as those from two female and one male animal over the first 364 dpw as represented in NMDS plots using the ΘYC distance metric. The stress for this representation was 0.15. Samples collected between 0 and 9 dpw are in blue, those from 11 to 140 dpw are in green, those from 141–150 dpw are in red, and samples from 364 dpw are in black. Larger characters are closer in the unrepresented axis and smaller characters are further into the unrepresented axis. Animated versions of the NMDS plots for each animal and all points together are available as supplementary data.
Observing that the overall community had a significantly different structure during the late period compared with the early period, we attempted to identify the bacterial populations responsible for the shift. At the phylum level, there were no significant differences between the early and late periods (p > 0.05). Among the early and late samples, 70.54% of the sequences affiliated with members of the Bacteroidetes and 29.21% affiliated with the Firmicutes. Minor members of the community affiliated with the Actinobacteria (0.09%), Proteobacteria (0.02%), Tenericutes (0.10%), TM7 (0.02%), and the Verrucomicrobia (0.02%). To obtain a higher resolution characterization of the community, we identified those OTUs that had an average relative abundance greater than 0.5% across all samples (i.e., minimum average of 9.5 sequences observed per sample; n = 32 OTUs). We used a repeated measures paired group analysis of variance to identify those OTUs that were differentially represented between the early and late periods. We identified 11 OTUs whose relative abundance increased and 16 whose relative abundance decreased between the early and late periods (Table 1). Interestingly, some closely related OTUs had different profiles in their relative abundance during the early and late periods. For example, two members of the genus Lactobacillus (OTUs 12 and 23) decreased in abundance and one (OTU 9) increased in abundance. Similar results were observed for members of the family Porphyromonadaceae, which showed significant decreases (OTUs 5, 7 and 8) and increases (OTUs 6 and 13) in relative abundance between the early and late periods; three OTUs affiliated with the Porphyromonadaceae did not exhibit a significant difference in relative abundance between the early and late periods (OTUs 1, 4 and 14).
Table 1. The difference in average relative abundance of OTUs between the early and late periods.
| OTU | Taxonomy (level) | Early (%) | Late (%) | P-value | |
|---|---|---|---|---|---|
| 19 |
Clostridiales (order) |
-9.95 |
1.52 |
0.00 |
5.4E-16* |
| 37 |
Clostridiales (order) |
-2.40 |
1.25 |
0.24 |
3.3E-11* |
| 28 |
Firmicutes (phylum) |
-2.36 |
0.84 |
0.16 |
5.2E-11* |
| 44 |
Clostridiales (order) |
-2.09 |
0.83 |
0.20 |
5.5E-11* |
| 25 |
Clostridiales (order) |
-1.79 |
1.43 |
0.41 |
9.7E-17* |
| 18 |
Clostridiales (order) |
-1.66 |
1.62 |
0.51 |
4.4E-16* |
| 23 |
Lactobacillus (genus) |
-1.34 |
1.03 |
0.41 |
1.7E-05* |
| 12 |
Lactobacillus (genus) |
-1.31 |
1.96 |
0.79 |
2.0E-10* |
| 30 |
Clostridiales (order) |
-1.22 |
1.81 |
0.78 |
2.6E-09* |
| 24 |
Clostridiales (order) |
-1.11 |
0.86 |
0.40 |
2.5E-04* |
| 5 |
Porphyromonadaceae (family) |
-1.08 |
6.44 |
3.03 |
4.1E-24* |
| 29 |
Oscillibacter (genus) |
-0.84 |
1.18 |
0.66 |
8.5E-07* |
| 26 |
Ruminococcaceae (family) |
-0.66 |
0.91 |
0.57 |
2.3E-05* |
| 2 |
Bacteroides (genus) |
-0.64 |
10.54 |
6.77 |
3.6E-08* |
| 3 |
Alistipes (genus) |
-0.49 |
7.89 |
5.61 |
1.5E-04* |
| 35 |
Clostridiales (order) |
-0.30 |
0.84 |
0.68 |
1.1E-02* |
| 7 |
Porphyromonadaceae (family) |
-0.18 |
4.02 |
3.55 |
1.2E-02* |
| 8 |
Porphyromonadaceae (family) |
-0.15 |
3.57 |
3.22 |
3.8E-02* |
| 1 |
Porphyromonadaceae (family) |
-0.07 |
11.84 |
11.24 |
2.80E-01 |
| 22 |
Blautia (genus) |
-0.05 |
1.60 |
1.54 |
7.90E-01 |
| 4 |
Porphyromonadaceae (family) |
-0.03 |
8.53 |
8.35 |
5.50E-01 |
| 14 |
Porphyromonadaceae (family) |
0.03 |
2.23 |
2.28 |
6.40E-01 |
| 16 |
Bacteroidetes (phylum) |
0.15 |
0.82 |
0.92 |
4.50E-02 |
| 21 |
Clostridiales (order) |
0.53 |
0.97 |
1.40 |
5.2E-05* |
| 6 |
Porphyromonadaceae (family) |
0.86 |
4.41 |
8.00 |
3.3E-42* |
| 9 |
Lactobacillus (genus) |
1.19 |
1.54 |
3.52 |
8.9E-06* |
| 17 |
Clostridiales (order) |
1.19 |
0.37 |
0.85 |
4.4E-07* |
| 11 |
Bacteroidales (order) |
1.50 |
1.12 |
3.15 |
6.0E-55* |
| 15 |
Barnesiella (genus) |
1.56 |
2.57 |
7.57 |
8.1E-51* |
| 13 |
Porphyromonadaceae (family) |
2.12 |
0.71 |
3.09 |
7.7E-53* |
| 10 |
Bacteroidales (order) |
2.21 |
1.17 |
5.41 |
1.3E-76* |
| 27 |
Bacteroidales (order) |
3.59 |
0.14 |
1.74 |
1.5E-46* |
| 20 | Turicibacter (genus) | 3.69 | 0.10 | 1.26 | 1.5E-24* |
Shaded cells represent OTUs that were classified as belonging to the Firmicutes and the non-shaded cells represent those belonging to the Bacteroidetes. Significance was assessed by using a repeated measure paired treatment analysis of variance for each OTU and correcting for multiple comparisons using an experiment-wise error rate of 0.05.34
Considering the clear shift in community structure between the early and late periods, we were curious whether the bacterial populations we observed early were the same as those found late or whether they were the same populations but in different abundance. Traditional metrics of membership (e.g., Jaccard β-diversity distance) are adversely affected by under sampling because rare populations are unevenly represented. To circumvent this difficulty, we assumed that the ten days within each period represented the core-microbiome for the entire period and for that mouse and calculated the fraction of sequences from the early and late periods that affiliated with a shared OTU for each mouse. Across the 12 mice, between 94.1 and 98.5% of the sequences from the early or late periods belonged to OTUs that were found in both periods and there was not a significant difference between the early and late periods (paired T-test; p = 0.67). When we looked for OTUs with an average relative abundance greater than 0.2% that were unique to either periods, we were unable to detect any that were unique to a period across all of the animals. These results suggest that although the membership of the early and late periods was largely the same, it was changes in the relative abundance of these populations that explained the observed differences in community structure.
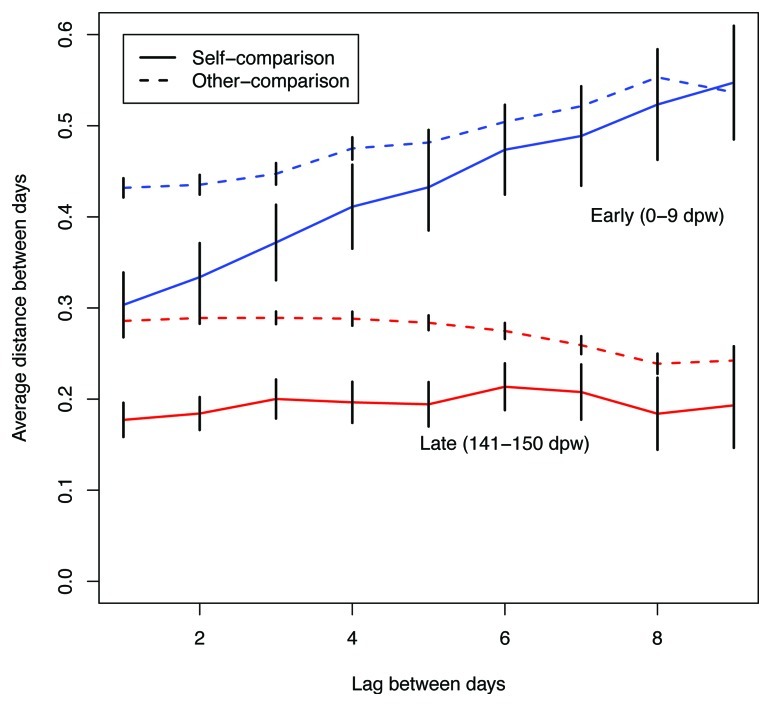
To measure the difference in stability among the early and late communities, we measured the community-level variation within each period for each animal by calculating the mean squared distance between all points within a period for each animal. Across the 12 animals the mean squared distances for each animal decreased by between 1.5 and 7.6-fold (p = 0.0001). To investigate this reduced variation further, we measured the association between varying sized time intervals for each mouse in the early and late periods (Fig. 3). The late samples had a lower level of variation between time points within the same animal and between different animals compared with the early samples. In addition, we observed that as the time interval between samples increased, the dissimilarity between samples increased for the early time points, while it remained constant for the late time points. These data suggest that the intra-animal variation was random within the early and late time periods. Further support for random temporal variation within a period was provided by non-significant co-occurence statistics, which indicated that the microbial populations were not associating in a preferential manner (C-score, Checkerboard, and V-ratio, all p > 0.05). We calculated the coefficient of variation (i.e., the ratio of the standard deviation to the mean relative abundance) within a period for each OTU and animal to identify those OTUs that exhibited differential stability between the two periods (Table 2). Six OTUs had a greater than 2-fold reduction in their coefficient of variation including three members of the order Bacteroidales (OTUs 10, 11 and 27), one member of the genus Turicibacter (OTU 20), one member of the genus Barnesiella (OTU 15), and one member of the family Porphyromonadaceae (OTU 13); these were also the 6 OTUs that had the most dramatic increases in relative abundance (Table 1). None of the OTUs had a significant increase in its coefficient of variation between the early and late periods.
Figure 3. Average ΘYC distances between days for the same animal (self-comparison) and between different animals (other-comparison) for 0 to 9 dpw (blue) and 141 and 151 (red) dpw. Error bars represent the 95% confidence interval.
Table 2. The difference between the coefficient of variation for the early and late periods for each OTU.
| OTU | Taxonomy (level) | Early | Late | P-value | |
|---|---|---|---|---|---|
| 27 |
Bacteroidales (order) |
-2.48 |
3.416 |
0.612 |
3.6E-07* |
| 10 |
Bacteroidales (order) |
-2.46 |
1.246 |
0.226 |
1.1E-04* |
| 11 |
Bacteroidales (order) |
-1.63 |
0.753 |
0.243 |
6.3E-05* |
| 20 |
Turicibacter (genus) |
-1.59 |
2.632 |
0.877 |
6.6E-06* |
| 15 |
Barnesiella (genus) |
-1.23 |
0.737 |
0.315 |
9.5E-05* |
| 13 |
Porphyromonadaceae (family) |
-1.18 |
0.956 |
0.421 |
1.2E-04* |
| 1 |
Porphyromonadaceae (family) |
-0.98 |
0.517 |
0.263 |
8.3E-05* |
| 6 |
Porphyromonadaceae (family) |
-0.93 |
0.519 |
0.273 |
1.1E-05* |
| 2 |
Bacteroides (genus) |
-0.79 |
0.700 |
0.405 |
4.5E-03* |
| 28 |
Firmicutes (phylum) |
-0.77 |
1.335 |
0.781 |
6.3E-02 |
| 16 |
Bacteroidetes (phylum) |
-0.76 |
0.539 |
0.319 |
6.7E-05* |
| 4 |
Porphyromonadaceae (family) |
-0.68 |
0.397 |
0.247 |
9.9E-05* |
| 30 |
Clostridiales (order) |
-0.61 |
1.055 |
0.689 |
2.5E-03* |
| 17 |
Clostridiales (order) |
-0.60 |
1.716 |
1.136 |
1.6E-02* |
| 24 |
Clostridiales (order) |
-0.51 |
1.716 |
1.209 |
1.6E-02* |
| 35 |
Clostridiales (order) |
-0.49 |
0.772 |
0.549 |
2.0E-04* |
| 7 |
Porphyromonadaceae (family) |
-0.49 |
0.536 |
0.382 |
1.6E-04* |
| 14 |
Porphyromonadaceae (family) |
-0.47 |
0.555 |
0.401 |
1.4E-05* |
| 8 |
Porphyromonadaceae (family) |
-0.42 |
0.553 |
0.415 |
1.2E-04* |
| 22 |
Blautia (genus) |
-0.36 |
1.156 |
0.898 |
1.1E-02* |
| 23 |
Lactobacillus (genus) |
-0.31 |
1.487 |
1.197 |
1.0E-01 |
| 9 |
Lactobacillus (genus) |
-0.31 |
1.575 |
1.269 |
4.4E-04* |
| 3 |
Alistipes (genus) |
-0.28 |
0.849 |
0.697 |
2.1E-01 |
| 37 |
Clostridiales (order) |
-0.27 |
1.350 |
1.116 |
1.0E-01 |
| 18 |
Clostridiales (order) |
-0.23 |
0.891 |
0.758 |
2.1E-02* |
| 21 |
Clostridiales (order) |
-0.22 |
0.798 |
0.687 |
4.3E-02 |
| 26 |
Ruminococcaceae (family) |
-0.21 |
0.895 |
0.773 |
3.1E-01 |
| 44 |
Clostridiales (order) |
-0.08 |
1.324 |
1.249 |
4.7E-01 |
| 12 |
Lactobacillus (genus) |
0.02 |
0.972 |
0.985 |
7.1E-01 |
| 29 |
Oscillibacter (genus) |
0.14 |
0.946 |
1.042 |
8.9E-01 |
| 25 |
Clostridiales (order) |
0.17 |
0.915 |
1.030 |
6.9E-01 |
| 5 |
Porphyromonadaceae (family) |
0.36 |
0.505 |
0.647 |
2.9E-01 |
| 19 | Clostridiales (order) | 1.67 | 1.447 | 4.606 | NA |
Shaded cells represent OTUs that were classified as belonging to the Firmicutes and the non-shaded cells represent those belonging to the Bacteroidetes. Significance was assessed by using a paired T-test for each OTU and correcting for multiple comparisons using an experiment-wise error rate of 0.05.34NA, because OTU19 was only observed in one animal among the late time points, it was not possible to perform the test. The numbers presented are for M005.
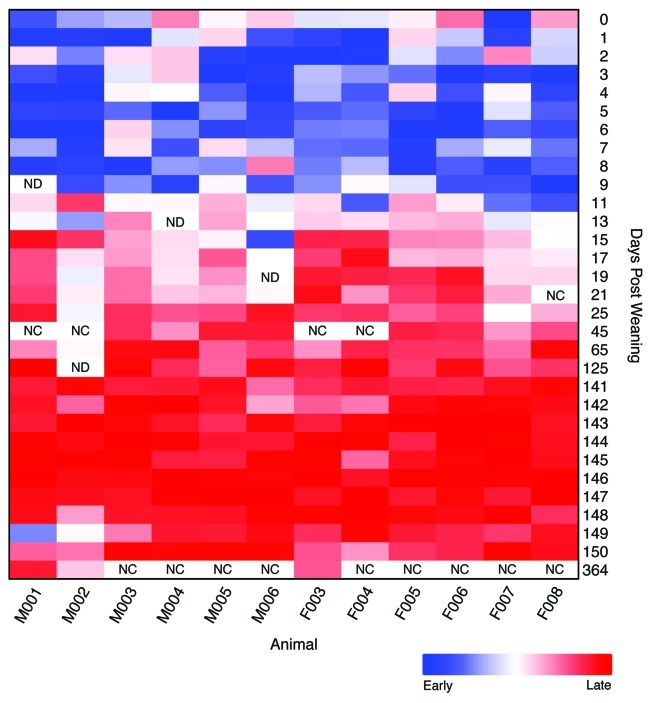
We performed additional sequencing using samples collected between 11 and 25 dpw and at 45, 65, and 125 dpw to better define the shift between the early and late periods. We calculated the root mean squared θYC distance (RMSD) between all of the samples for each animal to their early and late points for each mouse to determine when the community structure changed. The difference between the RMSD for each sample to the early and late time points indicated the affiliation of each sample to the early or late period. These data indicate that the communities all shifted to the late community structure by 11 to 17 dpw (Fig. 4). We also obtained sequence data at 364 dpw for three of the mice to quantify the long-term stability I the community. The community structures observed at 364 dpw clearly clustered among the late samples indicating that the late community structure exhibited constancy over long periods of time (Figs. 2 and 4).
Figure 4. Relative difference in root mean squared θYC distances to the early (0–9 dpw) and late (141–150 dpw) samples for each animal. Rectangles with “NC” indicate that the samples were not characterized and those with “ND” indicate those samples that had fewer than 1,900 sequences.
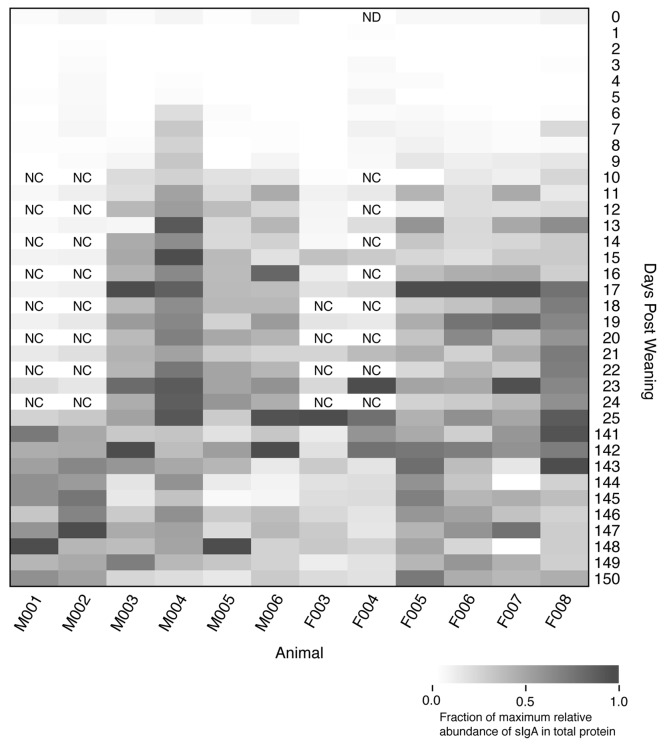
Prior to weaning the mice consumed solid chow and milk from their mother, which contained nutrients and high levels of IgA. We reasoned that although the behavioral and dietary effects of weaning would affect the microbiome, the dynamics of IgA in the weaned animals could explain a portion of the changes we observed in the microbiome. We quantified the amount of secretory IgA (sIgA) as a fraction of total protein using the same feces that were characterized by 16S rRNA gene sequencing. Interestingly, the sIgA levels increased an average of 25-fold between the early and late periods and were elevated by 6–11 dpw (Fig. 5). Although there was only a qualitative association between shifts in the community structure and sIgA levels, it appeared that the shift in sIgA preceded the shift in the community structure providing evidence to support the hypothesis that maturation of the immune system affected the observed changes in the microbiome.
Figure 5. Relative percentage of secretory IgA as total protein in the feces of each animal over the first 150 dpw. Rectangles with “NC” indicate that the samples were not characterized and the sample with “ND” did not yield useable data.
Discussion
The murine gut microbiome exists in two states in the 20 d after weaning. In addition, the second state exhibits greater stability than that observed immediately after weaning and the increased stability was not associated with a significant change in diversity. At least two possible explanations can account for this result. First, ecological mechanisms (e.g., competition) could alter the structure of the microbiome after the change in nutrients experienced at weaning. Second, as the immune system of the animal matures, for example with the replacement of maternal IgA with endogenous IgA, this could also provide a mechanism that drives change in microbiome structure. In addition, it is surprising that intra- and inter-animal variation was quite large given our control for the microbial inoculum (i.e., the mothers), diet, genetics, environment, sex, and age. Such variation and lack of clear successional patterns within a state suggest that in addition to ecological and immunological mechanisms, stochastic factors have a significant role in shaping the structure of the microbial communities within each state.
There also appears to be intra-genus specialization within the murine gut microbiome. As indicated in Tables 1 and 2, there were OTUs that affiliated with the same taxonomic lineage that behaved differently in the early and late periods. This result suggests that those populations interact differently with other members of the community or the host. Furthermore, it underscores the value of the OTU-based approach, which can provide greater taxonomic resolution than classification-based methods. A purely classification-based method would have lumped numerous OTUs into the same taxonomic grouping and effectively muted much of the signal that allowed us to explain the differences between the early and late periods. Further metagenomic-based analysis is needed to correlate intra-genus taxonomic variation with intra-genus functional variation.
Our study raises the interesting question of whether different community states are inherently more or less stable than others. By modulating the membership and structure of the microbiome, it is possible that the mice could experience different community states and allow us to probe diversity-stability relationships. Improved understanding of the community-based and host-based mechanism of stability will improve our understanding of the mechanisms of colonization resistance to pathogens and probiotics.
Materials and Methods
Animals and animal care
Two litters of C57BL/6 inbred mice (n = 4 and 8) were used in this study. The litters were obtained from a breeding colony established at the University of Michigan in 1991 using founder parents obtained from Jackson Laboratories. All mice were weaned at 21 d after birth and observations for 0 d post weaning were collected as the animal was transferred to its new cage. We obtained fresh fecal pellets as they were excreted and animal weights daily for each animal. All fecal pellets were stored at -80°C. Mice were housed in cages with autoclaved food, corncob bedding, and water. Cage changes occurred every 14 d and were performed in a laminar flow hood. All animal protocols used during this study were reviewed an approved by the University Committee on Use and Care of Animals at the University of Michigan.
Measurement of sIgA and total protein
We measured the concentration sIgA concentration using a mouse IgA ELISA quantitation kit with a standard curve (Bethyl). The total protein concentration was quantified using a protein quantification kit (BioRad) with a standard curve. sIgA levels were converted to a percentage of total protein and presented as a fraction of the maximum percent sIgA for each animal.
DNA extraction, amplification and sequencing
The remaining aliquot of suspended feces was used to extract and sequence 16S rRNA gene sequences from the microbial communities. Microbial genomic DNAs were extracted using the Roche MagNA Pure nucleic isolation kit on the MagNA Pure workstation. The V35 region was amplified and sequenced in the reverse direction as described elsewhere (http://www.hmpdacc.org/doc/HMP_MDG_454_16S_Protocol_V4_2_102109.pdf). Barcoded PCR primers allowed us to sequence 96 samples in parallel. We utilized 653 barcodes on seven half-picotiter plate 454 Titanium sequencing runs; the other half of the plate did not contain 16S rRNA gene fragments to avoid contamination between regions. We attempted to amplify and sequence the 16S rRNA gene from 29 or 30 fecal samples from each of the 12 mice (Fig. 3). All of the early and late time point samples for an animal were processed on the same sequencing run. This allowed us to limit the risks of batch effects that could cause us to observe differences between periods due solely to when the sequencing was performed. We also re-sequenced a mock community consisting of 21 pooled genomic DNA samples on three of the sequencing runs to assess the overall PCR and sequencing error rate.21
We successfully obtained sequence data from 643 of the barcodes; one of the missing barcodes represented the only sequencing data for that sample and the other missing barcodes were technical replicates. After curating the sequence data, we obtained between 1 and 22,116 sequences (median = 4,300; IQR = 3,380–5,243) with a median length of 247 bp from the remaining barcoded samples. Uneven sampling would result in the inclusion of disproportionate numbers of spurious sequences and would have a biased effect on a number of our community metrics. Therefore, depending on the type of analysis, we either rarefied our data to 1,900 sequences or randomly selected 1,900 sequences from each sample. This depth was selected because it allowed us to maintain deep sequencing coverage. Although 31 barcoded samples had fewer than 1,900 sequences and were removed from further analysis, most of these barcodes were technical replicates and only caused us to exclude four additional fecal samples from the rest of the analysis. Our microbial community analysis was based on 354 fecal samples.
Sequence curation and analysis pipeline
The sequence analysis pipeline implemented here largely follows our previously reported methods as implemented in the mothur software package.21,22 Specifically, we used a flowgram denoising algorithm,23 alignment to the SILVA reference alignment using a NAST-based algorithm,24-26 filtering of sequences positions to insure that the sequences overlapped in the same alignment space, and pre-clustering to allow up to a 2-bp difference between sequences.21 Sequences that were flagged as potential chimeras using UCHIME27 or that were classified as Chloroplasts were culled from the sequence collection. Sequences were assigned to OTUs based on a 3% difference cutoff using average neighbor algorithm. After processing the mock community sequence data in parallel with the sequences from the fecal samples, we observed an average sequencing error rate of 0.0001 and an average residual chimera rate of 2.0% (n = 2,327–5,870 sequences). When the mock community data were rarefied to 1,900 sequences, we observed between 20.6, 22.0, and 23.7 OTUs; 18 OTUs were expected. To measure the technical variation due to PCR and sequencing errors and biases, we calculated the Yue and Clayton (θYC) distance between the 276 of the fecal samples that were sequenced 2 to 4 times. The median θYC distance between replicate samples was 0.049 (mean = 0.059; sd = 0.036; IQR = 0.036–0.073). Data from these technical replicates were averaged in subsequent analysis following sub-sampling or rarefaction.
All sequences were classified using a Bayesian kmer-based approach with an 80% confidence threshold using the RDP training set version 6 (http://sourceforge.net/projects/rdp-classifier).28 Using an expanded training based on the greengenes reference taxonomy did not improve the classification of the sequences.29 A classification for each OTU was obtained by determining which taxonomy had the majority consensus of sequences within the OTU.30 To calculate phylogenetic diversity and UniFrac statistics, we constructed neighbor joining phylogenetic trees using the non-heuristic algorithm implemented in clearcut.31 Beta-diversity using OTU-based data was calculated using the θYC distance metric because it provides an even weighting of abundant and rare OTUs.32 Co-occurrence statistics and significance values were calculated as described by Gotelli33 for sample list-based data. The sff binary files, primers, barcodes, and a spreadsheet of MIMARKS compliant metadata are available at http://www.mothur.org/transition.
Supplementary Material
Acknowledgments
Grants from the National Institutes for Health to PDS (R01HG005975, P30DK034933, and U19AI090871), VBY (R01DK070875, P30DK034933, and U19AI090871), and JFP (U54HG004973) supported this research. Funding was also provided to PDS by the National Science Foundation (award #0743432) and to JFP by the Alkek Foundation.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
These authors contributed equally to this work.
Previously published online: www.landesbioscience.com/journals/gutmicrobes/article/21008
References
- 1.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–4. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dunbar J, Eichorst SA, Gallegos-Graves LV, Silva S, Xie G, Hengartner NW, et al. Common bacterial responses in six ecosystems exposed to 10 years of elevated atmospheric carbon dioxide. Environ Microbiol. 2012;14:1145–58. doi: 10.1111/j.1462-2920.2011.02695.x. [DOI] [PubMed] [Google Scholar]
- 3.Werner JJ, Knights D, Garcia ML, Scalfone NB, Smith S, Yarasheski K, et al. Bacterial community structures are unique and resilient in full-scale bioenergy systems. Proc Natl Acad Sci U S A. 2011;108:4158–63. doi: 10.1073/pnas.1015676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol. 2011;9:279–90. doi: 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
- 5.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. MetaHIT Consortium Enterotypes of the human gut microbiome. Nature. 2011;473:174–80. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–8. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fernández A, Huang SY, Seston S, Xing J, Hickey R, Criddle C, et al. How stable is stable? Function versus community composition. Appl Environ Microbiol. 1999;65:3697–704. doi: 10.1128/aem.65.8.3697-3704.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ives AR, Carpenter SR. Stability and diversity of ecosystems. Science. 2007;317:58–62. doi: 10.1126/science.1133258. [DOI] [PubMed] [Google Scholar]
- 9.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4578–85. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caporaso JG, Lauber CL, Costello EK, Berg-Lyons D, Gonzalez A, Stombaugh J, et al. Moving pictures of the human microbiome. Genome Biol. 2011;12:R50. doi: 10.1186/gb-2011-12-5-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–7. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–3. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 13.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4554–61. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reeves AE, Theriot CM, Bergin IL, Huffnagle GB, Schloss PD, Young VB. The interplay between microbiome dynamics and pathogen dynamics in a murine model of Clostridium difficile Infection. Gut Microbes. 2011;2:145–58. doi: 10.4161/gmic.2.3.16333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213–23. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffmann C, Hill DA, Minkah N, Kirn T, Troy A, Artis D, et al. Community-wide response of the gut microbiota to enteropathogenic Citrobacter rodentium infection revealed by deep sequencing. Infect Immun. 2009;77:4668–78. doi: 10.1128/IAI.00493-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Antonopoulos DA, Huse SM, Morrison HG, Schmidt TM, Sogin ML, Young VB. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect Immun. 2009;77:2367–75. doi: 10.1128/IAI.01520-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Samuel BS, Gordon JI. A humanized gnotobiotic mouse model of host-archaeal-bacterial mutualism. Proc Natl Acad Sci U S A. 2006;103:10011–6. doi: 10.1073/pnas.0602187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faith JJ, McNulty NP, Rey FE, Gordon JI. Predicting a human gut microbiota’s response to diet in gnotobiotic mice. Science. 2011;333:101–4. doi: 10.1126/science.1206025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schloss PD, Gevers D, Westcott SL. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS One. 2011;6:e27310. doi: 10.1371/journal.pone.0027310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–41. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ. Removing noise from pyrosequenced amplicons. BMC Bioinformatics. 2011;12:38. doi: 10.1186/1471-2105-12-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–96. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeSantis TZ, Jr., Hugenholtz P, Keller K, Brodie EL, Larsen N, Piceno YM, et al. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 2006;34(Web Server issue):W394-9. doi: 10.1093/nar/gkl244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schloss PD. A high-throughput DNA sequence aligner for microbial ecology studies. PLoS One. 2009;4:e8230. doi: 10.1371/journal.pone.0008230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–7. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Werner JJ, Koren O, Hugenholtz P, DeSantis TZ, Walters WA, Caporaso JG, et al. Impact of training sets on classification of high-throughput bacterial 16s rRNA gene surveys. ISME J. 2012;6:94–103. doi: 10.1038/ismej.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schloss PD, Westcott SL. Assessing and improving methods used in operational taxonomic unit-based approaches for 16S rRNA gene sequence analysis. Appl Environ Microbiol. 2011;77:3219–26. doi: 10.1128/AEM.02810-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheneman L, Evans J, Foster JA. Clearcut: a fast implementation of relaxed neighbor joining. Bioinformatics. 2006;22:2823–4. doi: 10.1093/bioinformatics/btl478. [DOI] [PubMed] [Google Scholar]
- 32.Yue JC, Clayton MK. A similarity measure based on species proportions. Commun Stat Theor M. 2005;34:2123–31. doi: 10.1080/STA-200066418. [DOI] [Google Scholar]
- 33.Gotelli NJ. Null model analysis of species co-occurrence patterns. Ecology. 2000;81:2606–21. doi: 10.1890/0012-9658(2000)081[2606:NMAOSC]2.0.CO;2. [DOI] [Google Scholar]
- 34.Benjamini Y, Hochberg Y. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J Roy Stat Soc B Met. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.