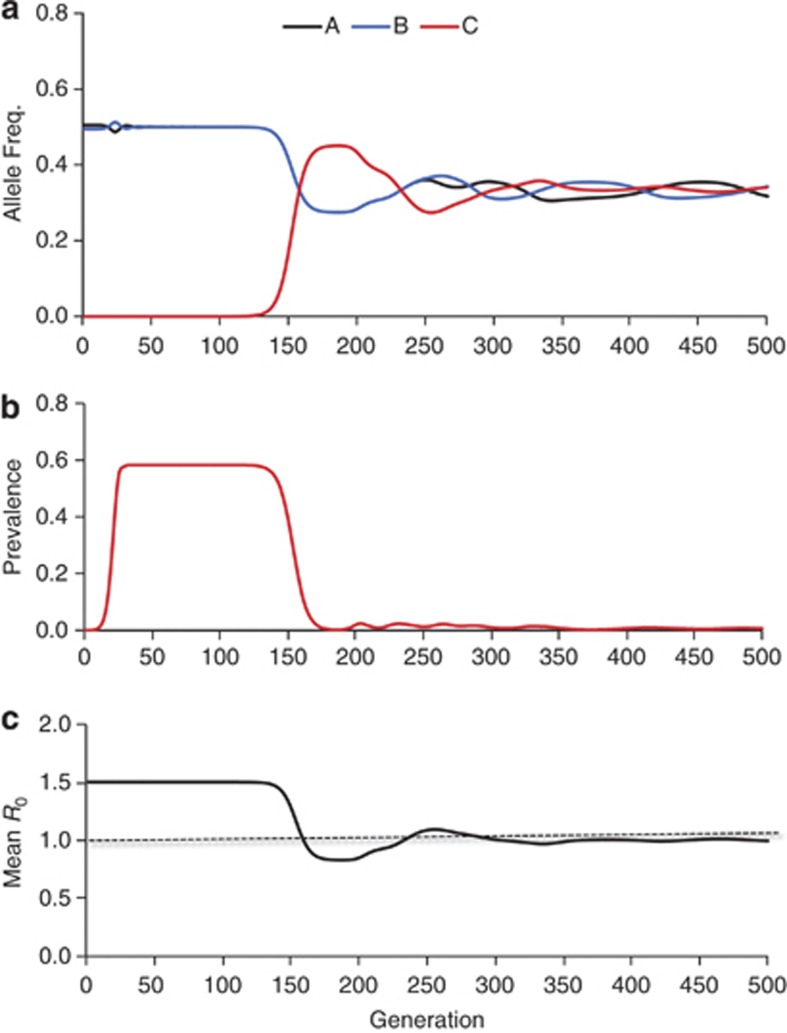
Figure 1.
The effect of increasing host genetic diversity on R0 and the prevalence of infection. A new allele was introduced at generation 100, which increased the number of possible haploid genotypes from six to nine. The simulation is based on the calculations in Lively (2010a); parameter values used were: ai=au=0.0001; bu=10; bi=7. As such, virulence was density independent and equal to 0.30 (=1−bi/bu; the parameter values are chosen only for the purpose of illustration of the main ideas). (a) Allele frequencies at the second locus, overtime, in which the new allele C was introduced at generation 100. Note the rapid spread of the newly introduced allele. (b) Prevalence of infection overtime. Note the dramatic decrease in infection prevalence following the spread of the new allele, C. (c) R0 overtime. Note the rapid decrease in R0 after the introduction of the new allele, C, at the second locus at generation 100. The dashed line denotes R0=1 (epidemiological threshold). Taken together, the results suggest that increasing genetic diversity by the introduction of a single novel allele in the host population can eliminate infection in tens of generations.