Abstract
The aim of this study was to examine the role of Programmed cell death 4 (Pdcd4) in colorectal adenocarcinoma (CRA). Pdcd4 expression was observed in both the nucleus and cytoplasm in colorectal adenocarcinoma, whereas Pdcd4 was expressed in the nucleus in normal colonic epithelial cells. Loss or weak expression of Pdcd4 was identified in 44 cases (40.7%) of cancer cells. Pdcd4 expression was associated with an increase in the nodal and clinical stage (p=0.022 and p=0.016, respectively). Nuclear staining was identified in 66 cases (61.15%), with no correlation with clinicopathological factors. Conversely, cytoplasmic staining for Pdcd4 was observed in 45 cases (41.7%), and increased according to nodal and clinical stage (p=0.011 and p=0.009, respectively), indicating that aberrant Pdcd4 expression leads to tumor progression. However, Pdcd4 expression was not correlated to disease-free survival time. This study demonstrated that during the tumorigenesis of CRA, loss of nuclear Pdcd4 expression occurs, and during tumor progression, aberrant cytoplasmic expression is present, suggesting a higher clinical stage. Although loss of Pdcd4 was not significantly correlated with survival time, as the prognosis of colorectal cancer varies depending on clinical stage including invasion depth, nodal status and metastatic status, cytoplasmic Pdcd4 expression may be a favorable prognostic marker in CRA.
Keywords: programmed cell death 4, colorectal adenocarcinoma
Introduction
Programmed cell death 4 (Pdcd4) is a ubiquitously expressed, novel tumor suppressor gene localized in chromosome 10q24, whose protein product plays a role in the suppression of tumorigenesis and tumor progression and invasion (1–4). Although Pdcd4 was first identified as being differentially upregulated during apoptosis, experimental evidence established it as a novel tumor suppressor (5–8). Previous studies indicated that the overexpression of Pdcd4 inhibited tumor promoter TPA-induced neoplastic transformation in JB6 and P-positive cells (4,9,10). Pdcd4 expression is often decreased in progressed carcinomas of the lung, ovary, breast, colon and prostate (7,11–13). Pdcd4 protein acts as an inhibitor that suppresses protein translation through its binding to and inhibition of the helicase activity of eukaryotic translation initiation factor 4A (eIF4A), the latter being a component of the protein translation complex (4,9,14). Certain studies investigating cellular functions of Pdcd4 demonstrated that it was capable of regulating molecules such as p21 (7), Cdk3, ornithine decarboxylase (1), carbonic anhydrase II (15) and JNK/c-Jun/AP-1 (6). It has also been shown to suppress the expression of the invasion-related urokinase receptor (u-PAR) gene, and to suppress invasion/intravasation via the Sp1/Sp3 promoter motifs in cancer (8). However, the physiological role of Pdcd4 is not clearly understood. The purpose of the present study was to examine the role of Pdcd4 in colorectal adenocarcinoma (CRA). The prognosis of colorectal cancer patients varies depending on the depth of the invasion of tumor cells and the lymph node status, and these factors are assessed only by a histopathological examination of surgically resected tissues (16). Pdcd4 expression and its subcellular localization were investigated using immunohistochemistry, and Pdcd4 expression was correlated with clinical and pathological parameters including histology, grade, stage and overall survival.
Materials and methods
Patients
Among patients who underwent curative surgery for CRA at Chosun University Hospital between January 1992 and December 2001, the present study was conducted on a non-consecutive series of 108 patients from whom paraffin-embedded tissues were obtained and relatively well preserved, medical records were complete and the patient status had been followed up. Patient survival was confirmed through telephone interviews and by mail. Patients who underwent preoperative chemoradiotherapy and emergency surgery, and patients who had evidence of hereditary non-polyposis colorectal cancer or familial adenomatous polyposis were excluded from the study.
The various clinicopathological parameters of the patients were confirmed by reviewing the patient medical records and pathology files. The correlation of clinicopathological parameters and immunohistochemical findings with survival was investigated for all 108 patients. Informed consent was obtained from each patient according to the institutional guidelines, and the research protocols were approved by the Ethics Committee of the University Hospital.
Histopathological analysis
Microscopic examination
Each tumor was re-evaluated by retrospective analysis of the medical records and the tissue slide files of the Department of Pathology. Age, gender, tumor size, histological subtype, degree of differentiation, depth of tumor invasion, status of lymph node metastasis and presence of distant metastasis were assessed. Stage was defined according to the TNM staging system of the American Joint Committee on Cancer (17). The examined tissues were fixed in 10% neutral formalin, and the prepared paraffin-embedded tissues were sectioned at a thickness of 4–5 μm. Hematoxylin and eosin staining was performed, and the sections were examined under a light microscope. A representative area suitable for the study purpose was selected, and slides were prepared for immunohistochemical analysis.
Immunohistochemical staining
Specimens in this study were tested using a rabbit polyclonal antibody against Pdcd4 (Abcam, Cambridge, MA, USA) according to the manufacturer’s instructions. Immunolocalization was performed using the mouse ImmunoCruz™ Staining System sc-2050 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), according to the manufacturer’s instructions. The staining process was performed according to a standard protocol. Briefly, the 4-μm sections that were obtained following formalin fixation and paraffin embedding were deparaffinized in xylene and rehydrated with distilled water through a graded series of ethanol solutions. The sections were then placed in a glass jar with 10 mM citrate buffer (pH 6.0) and were irradiated in a microwave oven for 15 min. The sections were allowed to cool in the jar at room temperature for 20 min. The slides were rinsed with Tris-buffered saline (TBS). A blocking reagent was added for 10 min after quenching the endogenous peroxidase activity in 0.3% hydrogen peroxide for 10 min. The slides were then washed as previously described, and the slides were subsequently subjected to the primary antibody reaction. Immunohistochemistry was performed using the Nexes ES (Ventana, Tucson, AZ, USA). Slides were incubated with the antibodies for 32 min. The Ventana basic DAB detection kit (catalog no. 760-001) was the secondary detection method. This kit includes biotinylated immunoglobulin secondary antibody, containing affinity-purified goat anti-mouse IgG and IgM (b200 lg/ml) and goat anti-rabbit IgG (b200 lg/ml) in phosphate buffer with preservative. Incubation was performed for 8 min, and was followed by conjugated streptavidin horseradish peroxidase. Slides were counterstained with hematoxylin (Ventana catalog no. 760-2021).
Analysis and interpretation of staining
Two pathologists, who were unaware of the clinical course of the subjects in order to exclude subjectivity, evaluated the results of the staining. Since Pdcd4 expression was identified in both the nucleus and the cytoplasm, nuclear and cytoplasmic staining was evaluated separately. Nuclear and cytoplasmic scoring was determined according to the percentage of positive nuclear staining and the intensity of cytoplasmic staining in cells, respectively (18). Nuclear scoring was as follows (18): score 1, negative; score 2, <30%; score 3, 30 to 70%; and score 4, >70%. Cytoplasmic scores were determined according to staining intensity: score 1, negative; score 2, weak staining intensity; score 3, intermediate staining intensity and score 4, strong staining intensity, in reference to a strongly stained tissue as a control. The overall Pdcd4 score was calculated by adding the nuclear and cytoplasmic scores, and 4 groups were defined: negative (total score, 1 and 2), weak staining (total score, 3 and 4), intermediate staining (total score, 5 and 6) and strong staining (total score, 7 and 8).
Statistical analysis
For the statistical analysis of Pdcd4 in CRA, its association with various clinicopathological factors, and survival rate or survival time, we used Pearson’s Chi-square test and linear-by-linear association to compare the categorical data. The life table and statistical significance were computed using the Kaplan-Meier method and the log-rank test. P<0.05 was considered to be statistically significant. The Stat View software package was used for all statistical analyses (Abacus Concepts, Berkeley, CA, USA).
Results
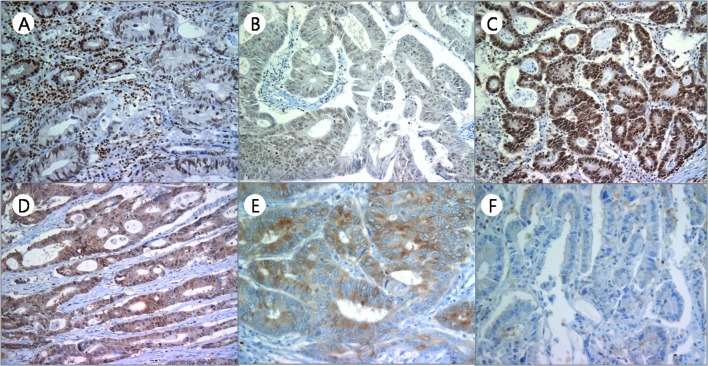
The clinical characteristics of the patients are shown in Table I. The average age at the time of surgery was 62.1 years and the ratio of male to female participants was 55:53 (50.9:49.1%). Tumors were mostly well- to moderately differentiated (88 cases, 81.5%), although 7 (6.5%) cases exhibited poor differentiation, and 13 (12.0%) cases were mucinous adenocarcinomas. Pdcd4 was ususally detected in the nuclei of non-neoplastic epithelial cells (Fig. 1A). However, in the cancer cells, Pdcd4 was observed in both the nuclei and the cytoplasms, and nuclear expression was downregulated. The nuclear, cytoplasmic and overall Pdcd4 scores were evaluated, respectively. An overall Pdcd4 score of 0, 1, 2 and 3 was identified in 11 (10.2%), 33 (30.5%), 57 (52.8%) and 7 (6.5%) cases, respectively. In contrast to normal epithelial cells, the majority of tumor cells had a weak to moderate cytoplasmic staining pattern for Pdcd4. A cytoplasmic expression score of 1, 2, 3 and 4 was observed in 63 (58.3%), 25 (23.1%), 17 (15.8%) and 3 (2.8%) cases, respectively (Table II). The cytoplasmic and overall score revealed a tendency to increase gradually according to the nodal status and AJCC clinical stage (linear-by-linear association, p<0.05). The cytoplasmic score also revealed significant difference among nodal status and AJCC clinical stages (Pearson’s Chi-square test, p<0.05). Moreover, nuclear expression scores of 1, 2, 3 and 4 were identified in 24 (22.2%), 18 (16.7%), 20 (18.5%) and 46 (42.6%) cases, respectively, and showed no tendency or significant difference with any of the clinicopathological parameters (Table II, Fig. 1B-F).
Table I.
Summary of clinicopathological factors.
| Characteristics | n (%) |
|---|---|
| Age | |
| ≤40 | 14 (13.0) |
| 51–59 | 28 (25.9) |
| 60–69 | 38 (35.2) |
| ≥70 | 28 (25.9) |
| Gender | |
| Male | 55 (50.9) |
| Female | 53 (49.1) |
| Pathological tumor classification (pT) | |
| pT1 | 2 (1.9) |
| pT2 | 21 (19.4) |
| pT3 | 80 (74.1) |
| pT4 | 5 (4.6) |
| Pathological lymph node classification | |
| pN0 | 73 (67.6) |
| pN1 | 26 (24.1) |
| pN2 | 9 (8.3) |
| Metastasis classification (M) | |
| M0 | 104 (96.3) |
| M1 | 4 (3.7) |
| AJCC classification | |
| I | 21 (19.4) |
| IIA | 49 (45.3) |
| IIIB | 2 (1.9) |
| IIIA | 2 (1.9) |
| IIIB | 22 (20.4) |
| IIIC | 8 (7.4) |
| IV | 4 (3.7) |
| Differentiation | |
| Well/Moderate | 88 (81.5) |
| Poor | 7 (6.5) |
| Mucinous | 13 (12.0) |
AJCC, American Joint Committee on Cancer.
Figure 1.
Various expression patterns of programmed death cell 4 (Pdcd4) in colorectal adenocarcinoma and normal cells. (A) Most normal colorectal epithelial cells revealed strong nuclear staining (upper left) compared with the staining in adenocarcinoma. (B-F) In carcinoma, various expression patterns were identified. (B) Moderate nuclear and weak cytoplasmic expression, (C) strong nuclear and weak cytoplasmic expression, (D) weak nuclear with moderate cytoplasmic expression, (E) extremely strong cytoplasmic expression, and (F) no expression in the either the nuclei or cytoplasms.
Table II.
Summary of Pdcd4 expression and correlation with clinicopathologic parameters.
| n | Pdcd4 expression | Nuclear expression | Cytoplasmic expression | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
||||||||||||||
| 0A | 1A | 2A | 3A | P-value | 1 | 2 | 3 | 4 | P-value | 1 | 2 | 3 | 4 | P-value | ||
| T stage | ||||||||||||||||
| 1 | 2 | 0 | 1 | 1 | 0 | 0.443 | 1 | 0 | 0 | 1 | 0.018d | 1 | 0 | 1 | 0 | 0.903 |
| 2 | 21 | 3 | 8 | 9 | 1 | 5 | 2 | 10 | 4 | 13 | 4 | 3 | 1 | |||
| 3 | 80 | 8 | 20 | 46 | 6 | 17 | 14 | 9 | 40 | 45 | 20 | 13 | 2 | |||
| 4 | 8 | 0 | 4 | 1 | 0 | 1 | 2 | 1 | 1 | 4 | 1 | 0 | 0 | |||
| N stage | ||||||||||||||||
| N0 | 73 | 10 | 24 | 36 | 3 | 0.133 | 17 | 12 | 14 | 30 | 0.961 | 49 | 16 | 6 | 2 | 0.011d |
| N1 | 35 | 1 | 9 | 21 | 4 | 7 | 6 | 16 | 16 | 14 | 9 | 11 | 1 | |||
| M stage | ||||||||||||||||
| M0 | 104 | 11 | 33 | 53 | 7 | 0.294 | 24 | 18 | 20 | 42 | 0.133 | 60 | 24 | 17 | 3 | 0.808 |
| M1 | 4 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 4 | 3 | 1 | 0 | 0 | |||
| Clinical stage | ||||||||||||||||
| Highd | 72 | 10 | 24 | 35 | 3 | 0.114 | 17 | 12 | 14 | 29 | 0.906 | 49 | 15 | 6 | 2 | 0.009d |
| Lowd | 36 | 1 | 9 | 22 | 4 | 7 | 6 | 6 | 17 | 14 | 10 | 11 | 1 | |||
| Diff.1 | ||||||||||||||||
| W/Ma | 88 | 11 | 26 | 46 | 5 | 0.359 | 22 | 14 | 15 | 37 | 0.724 | 54 | 17 | 15 | 2 | 0.440 |
| Pb | 7 | 0 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 3 | 1 | 0 | |||
| Mc | 13 | 0 | 3 | 9 | 1 | 1 | 2 | 4 | 6 | 6 | 5 | 1 | 1 | |||
| Diff.2 | ||||||||||||||||
| Non-M1 | 93 | 10 | 29 | 48 | 6 | 0.925 | 22 | 16 | 15 | 40 | 0.419 | 55 | 20 | 16 | 2 | 0.438 |
| M2 | 15 | 1 | 4 | 9 | 1 | 2 | 2 | 5 | 6 | 9 | 5 | 1 | 1 | |||
Pdcd4 expression: 0A, negative staining (score 1–2); 1A, weak staining (score 3–4); 2A, intermediate staining (score 5–6); 3A, strong staining (score 7–8). N stage - N0, N1. Clinical stage: High, clinical stage I and II; Low, clinical stage I and II. Diff1: W/Ma, well to moderate differentiation; Pb, poor differentiation; Mc, mucinous carcinoma. Diff2: Non-M1, non-mucinous carcinoma; M2, mucinous carcinoma.
Statistically significant, p<0.05.
Discussion
Pdcd4 was first identified as being differentially upregulated during apoptosis; however, experimental evidence has established it as a novel tumor suppressor (5–8). Pdcd4 has been identified as a suppressor of transformation (5,9,19), tumorigenesis, progression (1), invasion, matrix-metalloproteinase activation (3,8) and tumor growth (20). In the present study, the Pdcd4 expression pattern was investigated in CRA (Fig. 1). Nuclear expression exhibited a tendency to be lost or weak in tumor cells, whereas cytoplasmic expression was increased. The transfer of Pdcd4 between the nucleus and cytoplasm is understood to have a significant effect on the regulation of its function. Pdcd4 is localized predominantly in the nucleus under normal growth conditions but is exported to the cytoplasm upon serum withdrawal (14). A number of authors have studied the localization of Pdcd4 in various cell types. Yang et al (19) reported that in the mouse JB6 preneoplastic cell line, both nuclear and cytoplasmic localization of Pdcd4 were identified. In their study, Yoshinaga et al (21) found that Pdcd4 accumulated in the nucleus at the G0 phase of asynchronous cultures of human normal fibroblasts but was localized in the cytoplasm during the cell cycle in tumor cell lines. Goke et al (7) reported that 6 of 7 colon carcinomas examined revealed a complete absence of nuclear Pdcd4 staining and additional cytoplasmic staining in 4 tumors. According to this study, epithelial cells of the prostate, breast and lung exhibited intense nuclear but no cytoplasmic staining; a clear shift from nuclear localization to cytoplasmic staining was observed in all colonic adenomas investigated. In the case of invasive breast cancer tissues, both nuclear and cytoplasmic localization were observed. In an investigation conducted into colorectal cancer, in addition to a decreased expression level of Pdcd4, a significant loss of nuclear Pdcd4 from normal tissues to colonic adenomas and carcinomas was also observed (18), supporting the hypothesis that the intracellular localization of Pdcd4 plays a significant role in the regulation of tumor cell progression. In the present study, similar results to other authors were also demonstrated, in that a differential expression pattern of Pdcd4 was found between normal and carcinoma cells by IHC analysis. Additionally, the cytoplasmic expression level was shown to increase according to the AJCC clinical stage. We suggested that Pdcd4 translocates from the nucleus to the cytoplasm during colonic cancer development. As proposed by Zhang (23), the accumulation of Pdcd4 in the nuclei is crucial for apoptosis, and the regulatory mechanisms of the localization of Pdcd4 protein may play a significant role in the induction of apoptosis in hepatocellular carcinoma cells. Wei et al (13) speculated that the accumulation of Pdcd4 in the nucleus negatively regulates cell proliferation while the cytoplasmic sequestration of Pdcd4 may abolish its involvement in ovarian cancer cells. Palamarchuk et al (22) transfected 293 cells that stably expressed wild-type Pdcd4 and observed a primarily nuclear Pdcd4 staining pattern. However, an S457A mutant of Pdcd4 that was unable to be phosphorylated by Akt, in contrast to the wild-type, was not localized in the nucleus but in the cytoplasm, indicating the significance of this amino acid and Akt phosphorylation site in the localization of Pdcd4. However, this phenomenon requires further confirmation using a larger patient cohort, and in vitro studies are necessary in order to understand its regulatory mechanism. In conclusion, both the localization and expression level of Pdcd4 may be potential indicators of colonic cancer progression.
The present study investigated the Pdcd4 expression pattern in CRA using IHC, and analyzed the correlation between Pdcd4 expression and clinical and pathological parameters, including histology, grade, stage and overall survival. In conclusion, in CRA, nuclear Pdcd4 expression was decreased and inversely aberrant cytoplasmic staining was identified, which was significantly increased according to the nodal status and AJCC clinical stage. We suggest that Pdcd4 translocates from the nucleus to cytoplasm in CRA progression, and that increased cytoplasmic expression of Pdcd4 is a potential prognostic marker in CRA. However, the regulatory mechanism of alteration of subcellular localization remains to be further investigated.
Acknowledgements
This study was supported by the National Research Foundation of Korea (NRF) Grant funded by the Ministry of Education, Science and Technology (MEST) through the Research Center for Resistant Cells (R-13-1003-09).
References
- 1.Jansen AP, Camalier CE, Colburn NH. Epidermal expression of the translation inhibitor programmed cell death 4 suppresses tumorigenesis. Cancer Res. 2005;65:6034–6041. doi: 10.1158/0008-5472.CAN-04-2119. [DOI] [PubMed] [Google Scholar]
- 2.Hilliard A, Hilliard B, Zheng SJ, et al. Translational regulation of autoimmune inflammation and lymphoma genesis by programmed cell death 4. J Immunol. 2006;177:8095–8102. doi: 10.4049/jimmunol.177.11.8095. [DOI] [PubMed] [Google Scholar]
- 3.Yang HS, Matthews CP, Clair T, et al. Tumorigenesis suppressor Pdcd4 down-regulates mitogen-activated protein kinase kinase kinase kinase 1 expression to suppress colon carcinoma cell invasion. Mol Cell Biol. 2006;26:1297–1306. doi: 10.1128/MCB.26.4.1297-1306.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soejima H, Miyoshi O, Yoshinaga H, et al. Assignment of the programmed cell death 4 gene (Pdcd4) to human chromosome band 10q24 by in situ hybridization. Cytogenet Cell Genet. 1999;87:113–114. doi: 10.1159/000015408. [DOI] [PubMed] [Google Scholar]
- 5.Cmarik JL, Min H, Hegamyer G, et al. Differentially expressed protein Pdcd4 inhibits tumor promoter-induced neoplastic transformation. Proc Natl Acad Sci USA. 1999;96:14037–14042. doi: 10.1073/pnas.96.24.14037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bitomsky N, Bohm M, Klempnauer KH. Transformation suppressor protein Pdcd4 interferes with JNK-mediated phosphorylation of c-Jun and recruitment of the coactivator p300 by c-Jun. Oncogene. 2004;23:7484–7493. doi: 10.1038/sj.onc.1208064. [DOI] [PubMed] [Google Scholar]
- 7.Goke R, Barth P, Schmidt A, Samans B, Lankat-Buttgereit B. Programmed cell death protein 4 suppresses CDK1/cdc2 via induction of p21(Waf1/Cip1) Am J Physiol Cell Physiol. 2004;287:1541–1546. doi: 10.1152/ajpcell.00025.2004. [DOI] [PubMed] [Google Scholar]
- 8.Leupold JH, Yang HS, Colburn NH, Asangani I, Post S, Allgayer H. Tumor suppressor Pdcd4 inhibits invasion/intravasation and regulates urokinase receptor (u-PAR) gene expression via Sp-transcription factors. Oncogene. 2007;26:4550–4562. doi: 10.1038/sj.onc.1210234. [DOI] [PubMed] [Google Scholar]
- 9.Yang HS, Jansen AP, Nair R, et al. A novel transformation suppressor, Pdcd4, inhibits AP-1 transactivation but not NF-κB or ODC transactivation. Oncogene. 2001;20:669–676. doi: 10.1038/sj.onc.1204137. [DOI] [PubMed] [Google Scholar]
- 10.Yang HS, Knies JL, Stark C, Colburn NH. Pdcd4 suppresses tumor phenotype in JB6 cells by inhibiting AP-1 transactivation. Oncogene. 2003;22:3712–3720. doi: 10.1038/sj.onc.1206433. [DOI] [PubMed] [Google Scholar]
- 11.Li T, Li D, Sha J, Sun P, Huang Y. MicroRNA-21 directly targets MARCKS and promotes apoptosis resistance and invasion in prostate cancer cells. Biochem Biophys Res Commun. 2001;383:280–285. doi: 10.1016/j.bbrc.2009.03.077. [DOI] [PubMed] [Google Scholar]
- 12.Wen YH, Shi X, Chiriboga L, Matsahashi S, Yee H, Afonja O. Alterations in the expression of Pdcd4 in ductal carcinoma of the breast. Oncol Rep. 2007;18:1387–1393. [PubMed] [Google Scholar]
- 13.Wei NA, Liu SS, Leung TH, et al. Loss of Programmed cell death 4 (Pdcd4) associates with the progression of ovarian cancer. Mol Cancer. 2009;8:70. doi: 10.1186/1476-4598-8-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bohm M, Sawicka K, Siebrasse JP, Brehmer-Fastnacht A, Peters R, Klempnauer KH. The transformation suppressor protein Pdcd4 shuttles between nucleus and cytoplasm and binds RNA. Oncogene. 2003;22:1905–1910. doi: 10.1038/sj.onc.1206710. [DOI] [PubMed] [Google Scholar]
- 15.Lankat-Buttgereit B, Gregel C, Knolle A, Hasilik A, Arnold R, Goke R. Pdcd4 inhibits growth of tumor cells by suppression of carbonic anhydrase type II. Mol Cell Endocrinol. 2004;214:149–153. doi: 10.1016/j.mce.2003.10.058. [DOI] [PubMed] [Google Scholar]
- 16.Park JK, Hong R, Kim KJ, Lee TB, Lim SC. Significance of p-STAT3 expression in human colorectal adenocarcinoma. Oncol Rep. 2008;20:597–604. [PubMed] [Google Scholar]
- 17.Greene FL, Page DL, Fleming ID, et al. AJCC Cancer Staging Manual. 6th edition. Springer-Verlag; New York: 2002. pp. 157–164. [Google Scholar]
- 18.Mudduluru G, Medved F, Grobholz R, et al. Loss of programmed cell death 4 expression marks adenoma-carcinoma transition, correlates inversely with phosphorylated protein kinase B, and is an independent prognostic factor in resected colorectal cancer. Cancer. 2007;110:1697–1707. doi: 10.1002/cncr.22983. [DOI] [PubMed] [Google Scholar]
- 19.Yang HS, Jansen AP, Komar AA, et al. The transformation suppressor Pdcd4 is a novel eukaryotic translation initiation factor 4A binding protein that inhibits translation. Mol Cell Biol. 2003;23:26–37. doi: 10.1128/MCB.23.1.26-37.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Y, Knosel T, Kristiansen G, et al. Loss of Pdcd4 expression in human lung cancer correlates with tumour progression and prognosis. J Pathol. 2003;200:640–646. doi: 10.1002/path.1378. [DOI] [PubMed] [Google Scholar]
- 21.Yoshinaga H, Matsuhashi S, Fujiyama C, Masaki Z. Novel human Pdcd4 (H731) gene expressed in proliferative cells is expressed in the small duct epithelial cells of the breast as revealed by an anti-H731 antibody. Pathol Int. 1999;49:1067–1077. doi: 10.1046/j.1440-1827.1999.00995.x. [DOI] [PubMed] [Google Scholar]
- 22.Palamarchuk A, Efanov A, Maximov V, Aqeilan RI, Croce CM, Pekarsky Y. Akt phosphorylates and regulates Pdcd4 tumor suppressor protein. Cancer Res. 2005;65:11282–11286. doi: 10.1158/0008-5472.CAN-05-3469. [DOI] [PubMed] [Google Scholar]
- 23.Zhang H, Ozaki I, Mizuta T, et al. Involvement of programmed cell death 4 in transforming growth factor-beta1-induced apoptosis in human hepatocellular carcinoma. Oncogene. 2005;25:6101–6112. doi: 10.1038/sj.onc.1209634. [DOI] [PubMed] [Google Scholar]