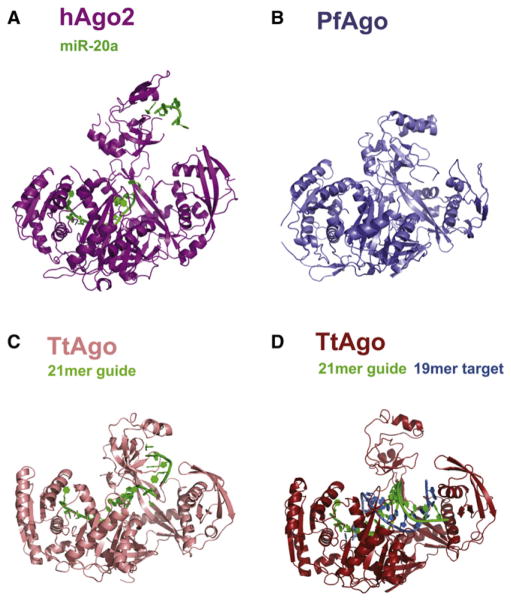
Figure 3. Structural Comparison of hAgo2, PfAgo, and Tt Ago.
Argonaute proteins across the three kingdoms of life show striking similarities in their overall structures. All structures were superimposed based on their respective PIWI domains and are oriented as in Figure 2. (A) hAgo2-miR-20a complex adopts the most open conformation of all full-length Argonaute structures determined to date. This is compared to the archaeal PfAgo (B), eubacterial TtAgo bound to a 21-mer DNA guide (C), and TtAgo in complex with a 21-mer DNA guide and a 19-mer RNA target (D). See also Figure S4.