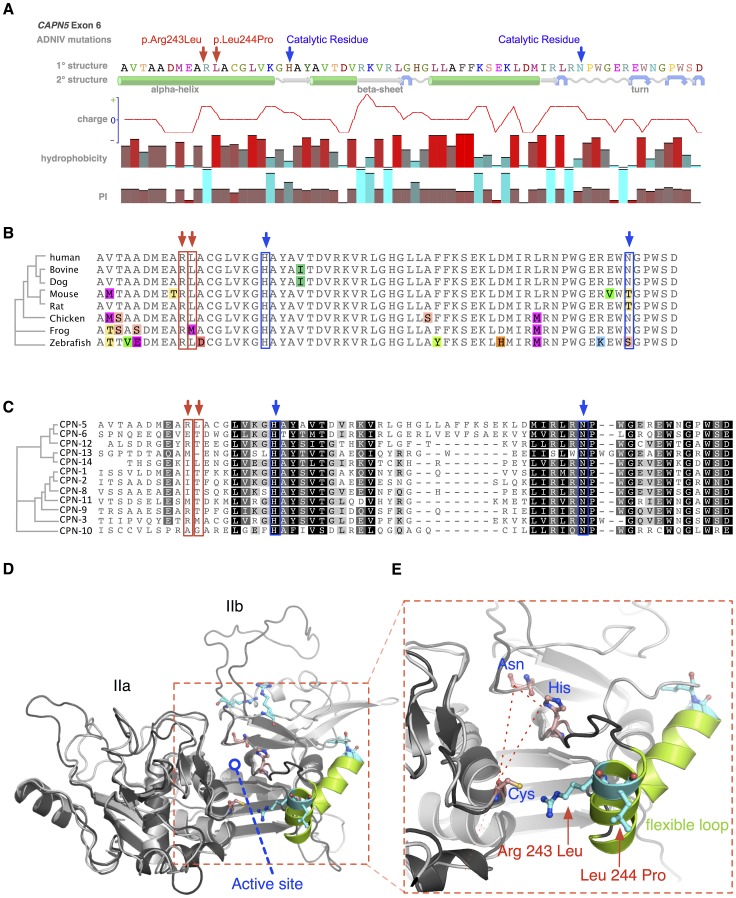
Figure 4. Protein structure modeling of calpain-5 and ADNIV mutants.
A. Both ADNIV mutations (red arrows) are located in exon 6, which encodes a portion of the catalytic domain and two of three catalytic residues (blue arrows). Primary protein sequence analysis shows the ADNIV-1/2 and ADNIV-3 mutations to be 8–9 amino acids upstream of the catalytic histidine residue. Secondary structure modeling shows that the two mutations are within a putative alpha helical domain. One mutated codon results in the loss of a basic residue, while the other introduces a proline into the putative alpha helix. B. Alignment of calpain-5 orthologs shows very high evolutionary conservation of the mutated residues (red arrows). Amino acid mismatches are color-highlighted. C. Twelve human calpain paralogs show significant differences in exon 6 (Black, 100% similarity; Dark grey, 80–100% similarity; Light grey, 60–80% similarity; White, less than 60% similarity). D. Three-dimensional modeling of the catalytic domain shows the location of the active site cleft (red outline). E. Magnified view of the active site cleft shows the catalytic triad (dashed line – blue text) and location of the two mutations (red arrows and text). Both mutations are located within a peptide loop that is homologous to a flexible loop of calpain-1 that undergoes a calcium-induced conformational change in association with regulation of the active site cleft (see text).