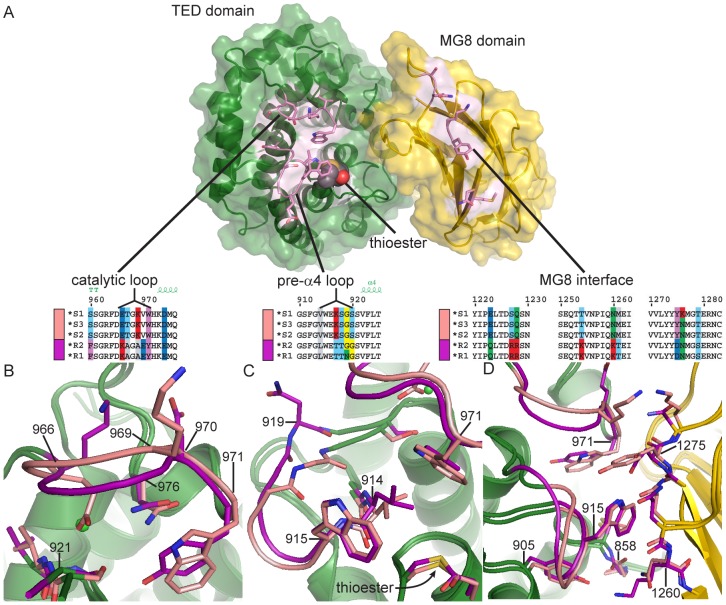
Figure 2. Comparison of TEP1*S1 and TEP1*R1 TED-MG8 interface.
(A) Exploded view of the TED-MG8 interface, the MG8 domain (yellow) has been rotated 90° with respect to the TED (green). The thioester bond is shown as VDW spheres, variable residues within the pre-α4 loop, catalytic loop and MG8 interface (pink) are shown as sticks. Sequence alignments for TEP1 alleles in each variable region are illustrated by superposition of TEP1*S1 (pink) and TEP1*R1 (magenta) structures in panels (B–D) with non-variable regions colored by domain for both alleles. (B) The catalytic loop (966–976); TEP1*S1 Glu 966 is directed towards Ser 921 in place of TEP1*R1 Tyr 971, causing a 2 Å displacement of residues 967–979. (C) The pre-α4 loop (914–920); TEP1*S1 Gly 919 is directed towards Val 914 whereas TEP1*R1 Asn 919 is directed towards the solvent, (3.9 Å displacement of Cα). (D) Complementary variation within the MG8 domain; TEP1*S1 Asn 1260 is within hydrogen bonding distance of Gly 858 but TEP1*R1 Lys 1260 also interacts with Tyr 884, and TEP1*S1 Tyr 1275 is not compatible with hydrogen bonding to Trp 915 as is the case for TEP1*R1 Asn 1275.