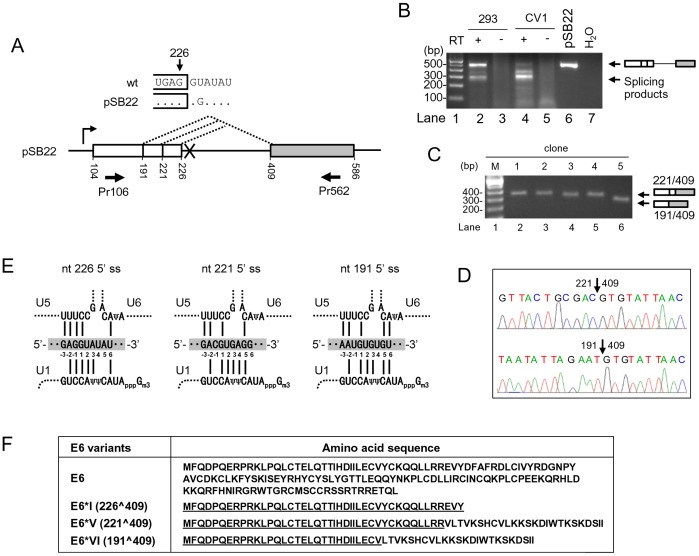
Figure 1. Identification of two novel alternative 5′ ss in the HPV16 intron 1.
(A) Diagram of alternative 5′ ss identified in this study by using pSB22, an HPV16 E6 expression vector containing a mutated nt 226 5′ ss (GU to GG, U to G at nt 228). Numbers below the pre-mRNA indicate nucleotide positions in the HPV16 genome. Boxes, exons; lines, introns; dashed lines, splicing directions. Arrows below the exons are a pair of primers used for RT-PCR. (B) RT-PCR result of alternative RNA splicing of E6 pre-mRNA in HEK293 or CV1 cells transfected with pSB22. RT-PCR products with a size of ∼300 bp were gel-purified and cloned. (C) Plasmid DNAs extracted from five bacterial colonies were digested by EcoRI and analyzed by agarose gel electrophoresis, showing the insert size. (D) Sequence chromatograms showing splice junctions of colonies 1–4 (nt 221/409) and the colony 5 (nt 191/409). (E) Illustration of the base pairing of three alternative 5′ ss (shaded, middle) in HPV16 intron 1 with U1, U5 and U6 snRNAs. Exon sequences are labeled as −1, −2 and −3 from the 5′ end of the intron. Base pairing is indicated with solid lines. Ψ, pseudo-uridine; Gm3, 2,2,7-tri-methyl guanosine. (F) Coding potentials of spliced E6E7 transcripts by using the nt 221 (E6*V) or nt 191 (E6*VI) 5′ ss. Amino acid residues underlined in E6*I, E6*II, E6*V and E6*VI are identical to the N-terminus of wt E6.