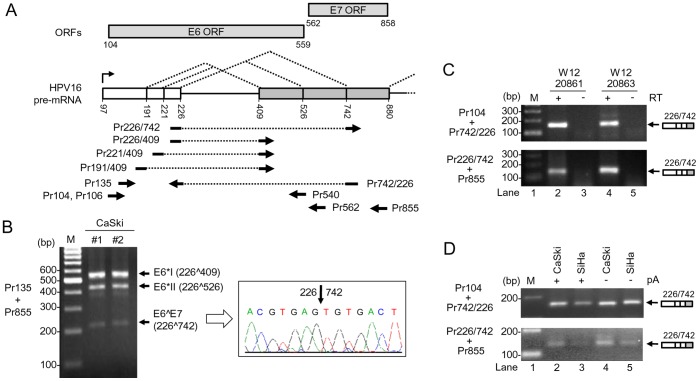
Figure 2. Utilization of nt 742 3′ ss in multiple HPV16+ cervical cancer cell lines.
(A) Diagram of splicing directions of HPV16 E6E7 pre-mRNA from three alternative 5′ ss at nt 191, 221, and 226 to three alternative 3′ ss at nt 409, 526 and 742. Numbers represent nucleotide positions in the HPV16 genome. Arrows under the E6E7 pre-mRNA structure indicate primers used in RT-PCR in this and all other figures. Rectangles above the E6E7 pre-mRNA structure indicate ORFs of E6 and E7. (B) Two preps of total RNA extracted from CaSki cells were analyzed by RT-PCR with a primer pair of Pr135 and Pr855 (left). The 214 bp RT-PCR product was purified from the gel and sequenced as a splicing product from nt 226 to 742 (right). (C) E6∧E7 expression is detectable by RT-PCR from W12 subclone cell lines 20861 and 20863 which harbor integrated and episomal HPV16 genome, respectively. (D) E6∧E7 expressed in HPV16+ CaSki and HPV16+ SiHa cells are polyadenylated. Total RNA with or without polyadenylation (pA) was used for RT-PCR with the indicated splice junction primers on the left (C and D).