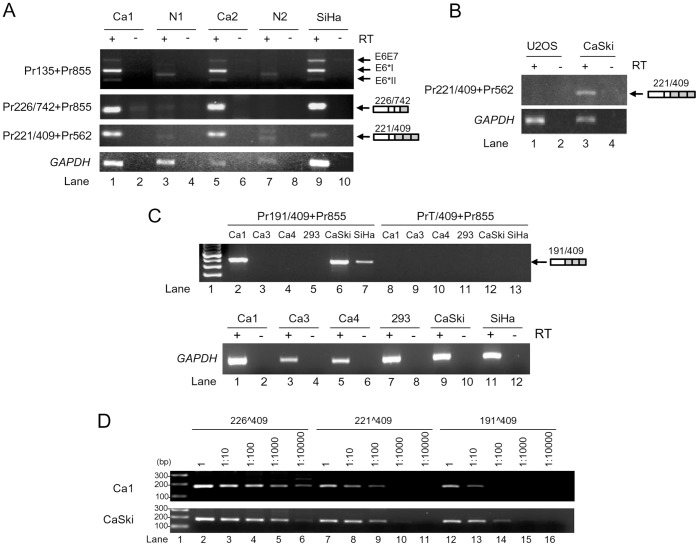
Figure 3. Usage of alternative 5′ ss and 3′ ss in HPV16+ cervical cancer tissues and cell lines.
(A) RT-PCR for total RNA from cervical tissues and SiHa cells with the indicated primer pairs (Fig. 2A) on the left, showing HPV16 splicing product from nt 221 5′ ss to nt 409 3′ ss in HPV16+ cervical cancer tissues (Ca) and SiHa cells. Expression of E6∧E7, E6*I, and E6*II served as positive controls. Normal cervical tissues (N) served as negative controls. A relative smaller or larger band in the normal tissue was a non-specific amplicon. (B) RT-PCR results for total RNA from HPV16+ CaSki cells and U2OS cells for splicing product from nt 221 to nt 409. HPV16-negative U2OS cells served as a negative control. (C) RT-PCR results of total RNA from HPV16+ cervical cancer tissues, CaSki, SiHa and HEK293 cells, showing the splicing product from nt 191 to nt 409. A 191∧409 splice junction 5′ primer and its 3′ half primer starting from nt 409 3′ ss were used in combination with a 3′ primer Pr855 for this comparison. HPV16-negative HEK293 RNA served as a negative control. GAPDH RNA served as a loading control. (D) Titration of the relative usage of nt 226, 221 and 191 5′ ss in cervical cancer tissues and CaSki cells by RT-PCR. The cDNAs of Ca1 and CaSki from panel C were serially diluted and amplified for the splicing products of 226∧409, 221∧409 and 191∧409 with each corresponding splice junction 5′ primer Pr226/409, Pr221/409, or Pr191/409 in combination of a common 3′ primer Pr562.