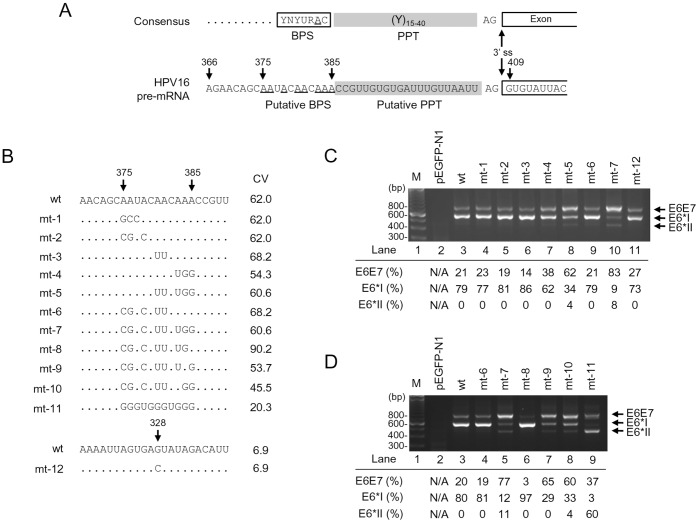
Figure 4. Mapping of the BPS in splicing of the HPV16 nt 409 3′ ss.
(A) Consensus splicing elements (3′ ss, BPS, polypyrimidine tract [PPT]) in a typical 3′ ss (upper) and their corresponding RNA sequences in the HPV16 nt 409 3′ ss (lower). (B) RNA sequences of putative wild type (wt) BPS and its derived mutants (mt 1–11) in HPV16 E6E7 constructs used in the study. Arrow heads indicate the nucleotide position in the HPV16 genome. Unchanged ribonucleotides in each mutant are indicated by dots. A mutant G328C BPS reported by another lab [46] was included in this study for verification. The nucleotide G at nt 328 position in the HPV16 genome was substituted with a C in the mutant construct (mt-12). CV, consensus value of the putative BPS [23]. (C and D) Identification of the nt 385A as a preferential branch site for splicing of the nt 409 3′ ss. RT-PCR results were obtained by using a primer pair of Pr106 and Pr855 (Fig. 2A) from total RNA extracted from HEK293 cells 24 h after transfection with a wt or mt construct as indicated above the agarose gel. Vector pEGFP-N1 served as a negative control (lane 2 in both C and D). M, size marker of 100-bp DNA ladders (lane 1 in both C and D). Identities of PCR products derived from unspliced E6E7 and spliced E6*I and E6*II RNA are shown on the right. Relative ratios (%) of the unspliced E6E7 (intron 1 retention) and spliced E6*I (nt 226∧409) and E6*II (nt 226∧526) RNA were calculated by the signal intensity of each product and shown below the gel.