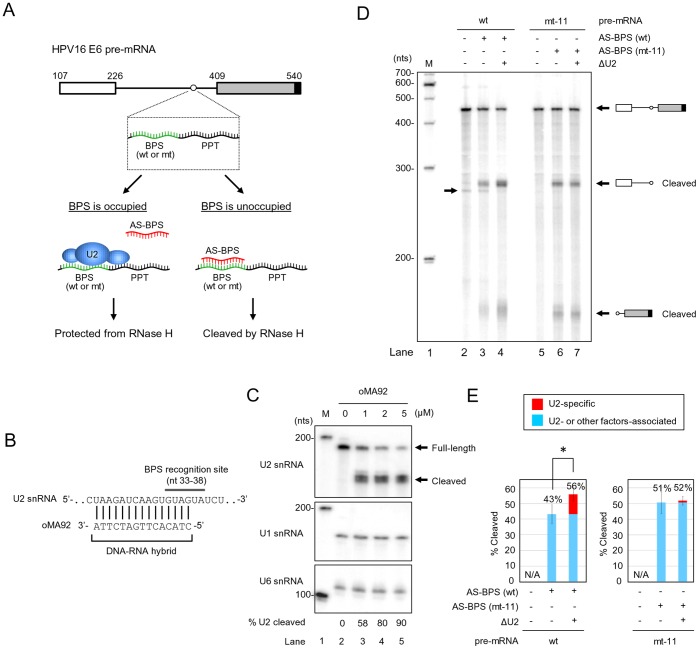
Figure 6. RNase H digestion to quantify U2 snRNP association with the wt or mt-11 BPS in the HPV16 intron 1.
(A) Flow chart of RNase H protection assays. A 32P-labeled HPV16 E6 RNA with a wt or mt-11 BPS was incubated with HeLa nuclear extract with or without U2 depletion in the presence of an antisense (AS) DNA oligo to wt or mt-11 BPS and RNase H. Association of U2 snRNA or other factors (unlabeled ovals) with the mapped BPS prevents the antisense DNA oligo to specifically access the correspondent region, thereby preventing RNase H cleavage of the testing pre-mRNA. Otherwise, binding of the antisense DNA oligo to the mapped BPS would form a DNA-RNA duplex and trigger RNase H digestion. (B) Diagram of DNA oligo-mediated U2 depletion from HeLa nuclear extract by RNase H digestion. A DNA oligo (oMA92) antisense to U2′s BPS recognition site was incubated at 30°C with HeLa nuclear extract for 30 min and the mixture was then digested by RNase H at 30°C for additional 10 min. (C) Determination of U2 depletion efficiency from HeLa nuclear extract by Northern blot. Total RNA from HeLa nuclear extract with or without U2 depletion by U2 DNA oligo-mediated RNase H digestion was quantified by Northern blot with a 32P-labeled DNA oligo specific for U2 (oKY50). U1 and U6 served as loading controls. U2 depletion efficiency was expressed as a ratio (%) of cleaved U2 vs remaining full-length U2 dividing by total signal intensity of U2 in each reaction condition of RNase H digestion. (D and E) U2 interaction with the mapped BPS prevents oligo-directed RNase H digestion. HeLa nuclear extracts with or without U2 depletion (ΔU2) were compared for DNA-oligo-mediated RNase H cleavage of HPV16 pre-mRNA with a wt or mt BPS (A). The reaction products were resolved in a 6% denaturing PAGE gel. Identities of RNase H cleavage products are shown on the right of the gel (D). An arrow on the left of the gel (D) indicates a spliced product from the wt pre-mRNA during 30 min incubation of RNase H digestion in the presence of HeLa nuclear extract. Cleavage efficiency (%) (E) of HPV16 E6 pre-mRNA with a wt or mt BPS from DNA oligo-mediated RNase H digestion in each reaction was calculated from four independent experiments by the sum of signal intensity of all cleavage products divided by the sum of signal intensity of both full-length pre-mRNA and cleavage products. *, P<0.05 by Student′s t-test.