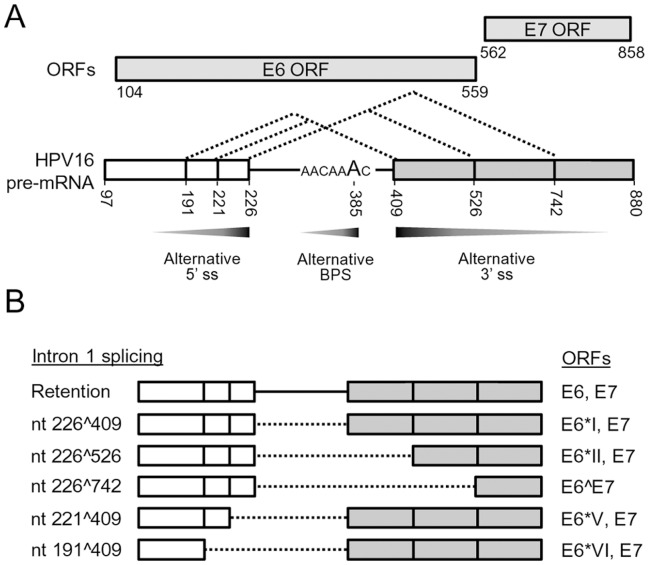
Figure 8. Summary of the characterized features of HPV16 E6E7 intron 1.
(A) Preferential selection of a proximal splice site and branch site in splicing of the HPV16 E6E7 intron 1 which contains three suboptimal 5′ ss (nt 191, 221 and 226), three suboptimal 3′ ss (nt 409, 526 and 742), and a cluster of branch site As from nt 383 to nt 385. In this regard, a proximal nt 226 5′ ss is preferentially selected over the nt 221 and nt 191 5′ ss, a proximal nt 409 3′ ss is efficiently selected over the nt 526 and nt 742 3′ ss, and the branch site A at nt 385 is preferentially utilized over the nt 384 A or 383 A, to minimize splicing energy and length of the excising intron 1. The preferential selection of an alternative 5′ ss, 3′ ss and branch site is exercised by the principle of proximity and is indicated by graded grey arrows for more (dark grey) to less (light grey) splicing of the HPV16 E6E7 intron 1. (B) Alternative splicing of HPV16 E6 intron 1 leads to produce multiple E6 RNA species detectable in cervical cancer tissues and their derived cell lines. Dash lines indicate the spliced intron 1 from each isoform of E6E7 RNA and vertical lines in white (exon 1) and grey boxes (exon 2) stand for splice sites. ORFs for splicing variants of viral E6 and E7 are indicated on the right.