FIGURE 3.
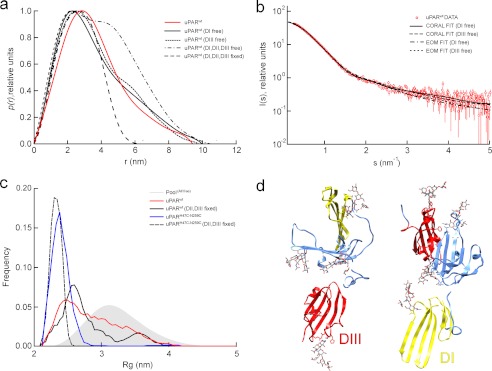
Rigid body modeling of the ligand-free uPAR variants. a, p(r) functions calculated for ensembles of rigid body models of uPAR where the interdomain orientation of either one or all three domains are randomized. The experimental p(r) function for uPARwt (solid red line) is approximately as compact as the structures, where only one of the domains is randomly orientated. b, fits of individual uPARwt models and ensembles to the SAXS data from CORAL rigid body refinement and EOM ensemble selection are shown, respectively, keeping two domains fixed in their relative orientation from the crystal structure and allowing one domain to be flexible. c, Rg distribution of the selected EOM ensembles. The closed mutant is best represented by a compact and relatively homogeneous ensemble, whereas the wild-type receptor is best fit by a heterogeneous ensemble. This suggests the presence of interdomain dynamics in the wild-type protein. Representative models resulting from rigid body modeling to the SAXS data are shown in d. The SAXS data fit well to models where either domain I or domain III is detached from the rest of the protein and projects away from the protein.