FIGURE 1.
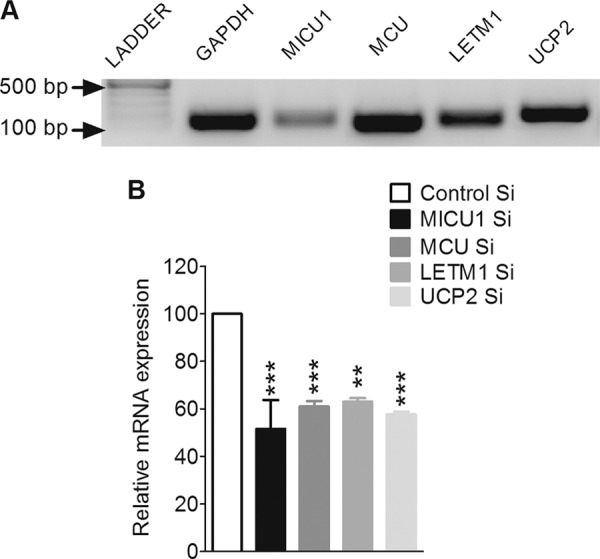
Expression analysis and silencing of MICU1, MCU, LETM1, and UCP2 in INS-1 832/13 cells. A, detection of mRNAs of MICU1, MCU, LETM1, and UCP2 was done by reverse transcription-polymerase chain reaction using a cDNA synthesis kit followed by an amplification with conventional PCR using gene-specific primers. Amplified PCR products were electrophoretically detected on a 1% agarose gel using a 100-bp ladder. GAPDH was used as an internal control. B, efficiency of MICU1 and MCU siRNAs was validated by quantitative real time PCR. Total RNA was isolated 48 or 72 h after transfection with siRNAs against MICU1 (n = 3), MCU (n = 4), LETM1 (n = 3), and UCP2 (n = 3). GAPDH was used as a housekeeping gene. Relative mRNA expression of each gene was compared with respective controls and shown as the percentage of maximum response. ***, p < 0.001, **, p < 0.01 versus respective controls. Control Si, control siRNA.
