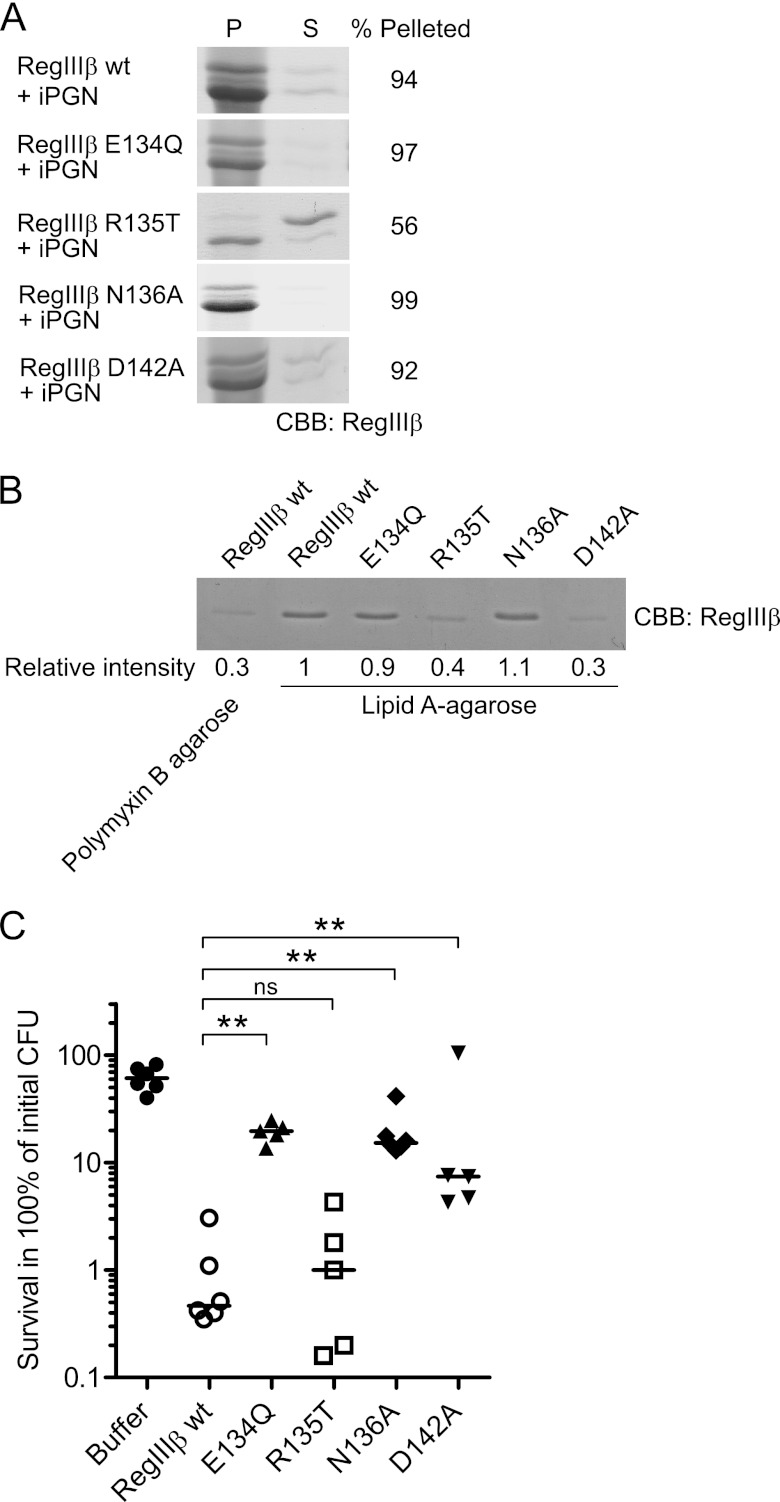
FIGURE 8.
Loop 2 is essential for substrate recognition of peptidoglycan and lipid A and for the bactericidal activity of RegIIIβ. A, peptidoglycan binding assays of the loop 2 mutants of RegIIIβ. 10 μg of RegIIIβ proteins, wild-type RegIIIβ (WT), E134Q, R135T, N136A, and D142A point mutants in loop 2 were added to 50 μg of insoluble peptidoglycan (iPGN) from B. subtilis. After incubation, the mixture was pelleted by centrifugation. The resulting pellet (P) and supernatant (S) fractions were analyzed by SDS-PAGE and CBB staining. Percent of band intensity of the pellet fraction was calculated using ImageJ software. B, lipid A binding assays of the loop 2 mutants of RegIIIβ. Purified wild-type RegIIIβ or point mutated RegIIIβ E134Q, R135T, N136A, or D142A were incubated with lipid A-immobilized polymyxin B-agarose. After washing the agarose, the bound RegIIIβ proteins were eluted by boiling in SDS-PAGE sample buffer and then subjected to SDS-PAGE and analyzed by CBB staining. The band intensity was calculated using ImageJ software. The relative intensity of wild-type RegIIIβ bound to the lipid A-immobilized agarose was defined as “1”. C, in vitro killing assay of the Loop 2 mutants of RegIIIβ. S. typhimurium wild-type strain (SL1344) from the middle logarithmic growth (A600 = 0.5) was incubated with purified recombinant RegIIIβ wild-type E134Q, R135T, N136A, or D142A (10 μm) for 30 min at 37 °C, and bacterial survival was quantified by dilution plating. Assays were performed in at least five independent experiments. The bar shows the median. **, p < 0.01; ns, not significant (Mann-Whitney U test).