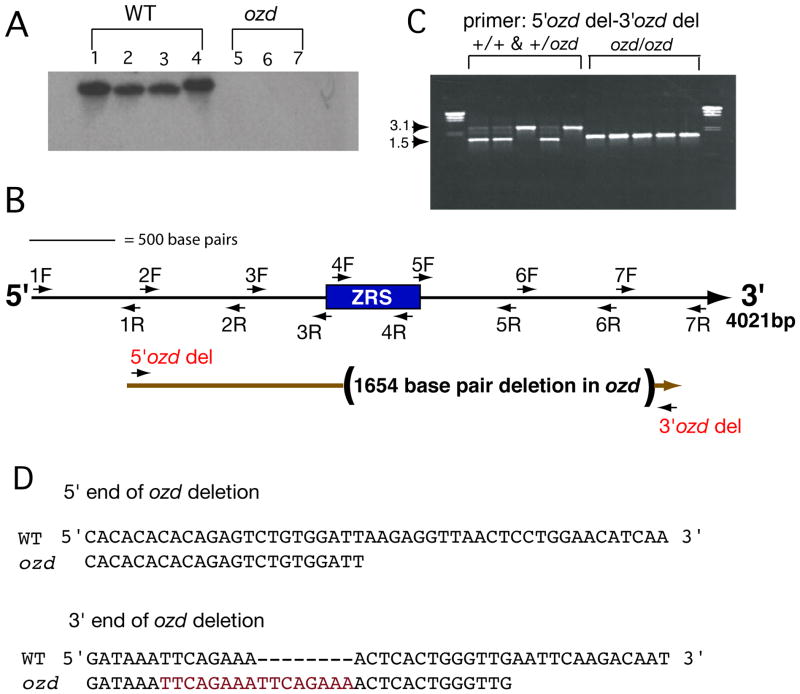
Figure 1. The chicken oligozeugodactyly (ozd) mutant contains a deletion of the ZRS.
(A) Genomic DNA from phenotypically normal (normal, +/+, and carriers, +/ozd, lanes 1–4) or known ozd mutants (lanes 5–7) was digested with SalI and BamHI. The resulting blot was probed with a 477 base pair ZRS fragment. A distinct band is present in all of the phenotypically normal lanes, but not in any ozd lane. (B) A BAC clone containing the conserved 477 base pair intron 5 region (rectangle) was obtained and ~4 kilobases of intron 5 was sequenced. Approximate locations of primers are shown as arrows, where the direction of the arrowhead depicts whether the primer was used to sequence (forward primers- arrows facing away from conserved region) or to amplify a PCR product from ozd samples (reverse primers- arrows facing towards conserved region). Primer pairs are numbered. The mutation in ozd was determined to be a 1654 base pair deletion within LMBR1 intron 5, deleting all but the first 135 base pairs of the ZRS and several hundred base pairs of intronic sequence (shown in parentheses). (C) 5′ozd del and 3′ozd del primers were used to amplify fragments from normal, heterozygous, and ozd genomic DNA samples. The normal band is ~3.1 kilobases and the mutant band is ~1.5 kilobases, for further explanation, see Results section of text. (D) Shown here are the 5′ and 3′ breakpoints in ozd mutants. Note the duplicated octamer (shown in red) near the 3′ breakpoint of the ozd deletion.