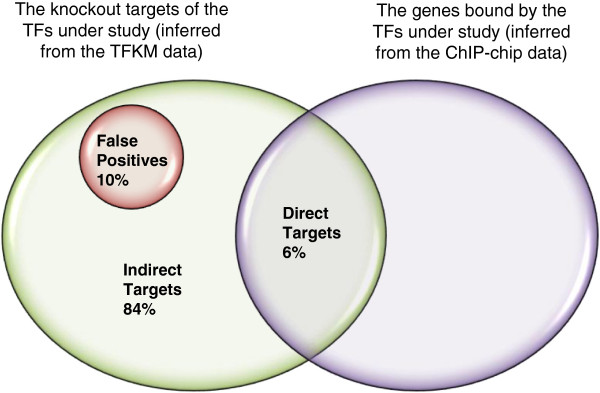
Figure 3.
The overlap between the TFKM data and the ChIP-chip data is very low. Simply intersecting the TFKM data with the ChIP-chip data can only interpret 6% of the original dataset, which corresponds to those biologically interpretable TF knockout targets with the shortest paths of length one (i.e., the direct targets of the knocked-out TF). Our method can further interpret the other 84% of the original dataset, which corresponds to those biologically interpretable TF knockout targets with the shortest paths of length larger than one (i.e., the indirect targets of the knocked-out TF). The other 10% of the original dataset, for which no path could be found, is regarded as the false positives.